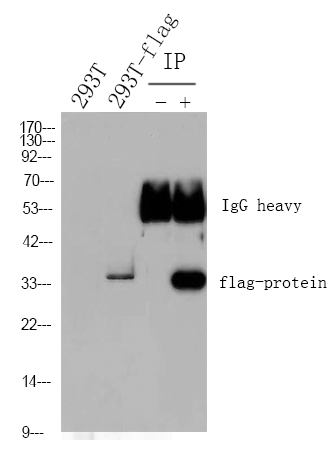
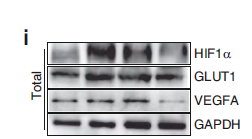
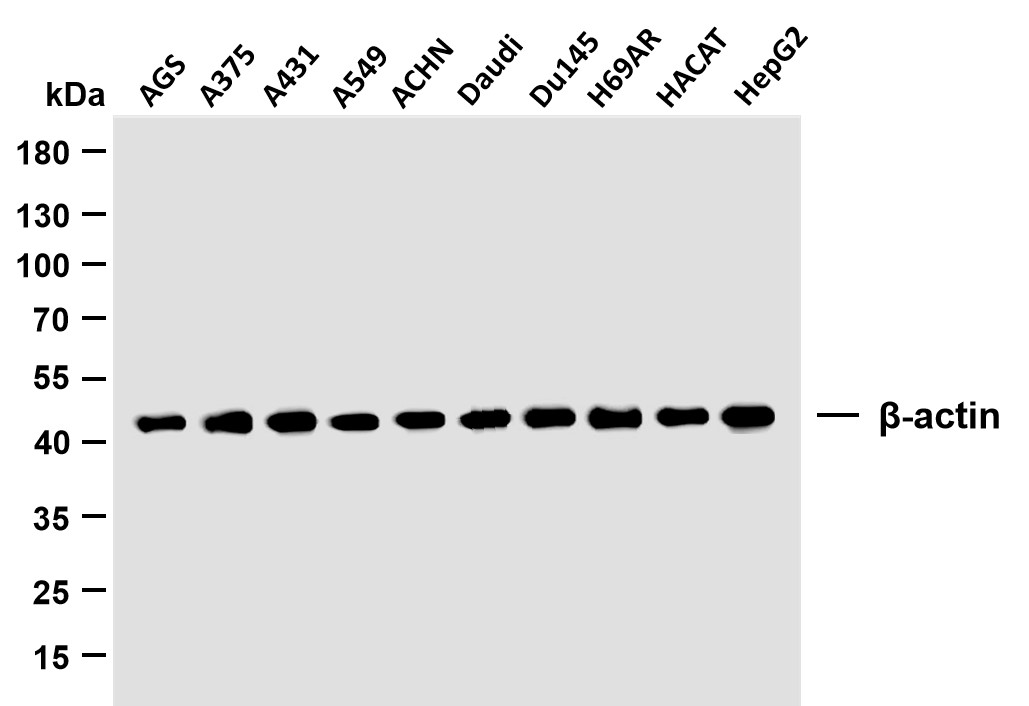
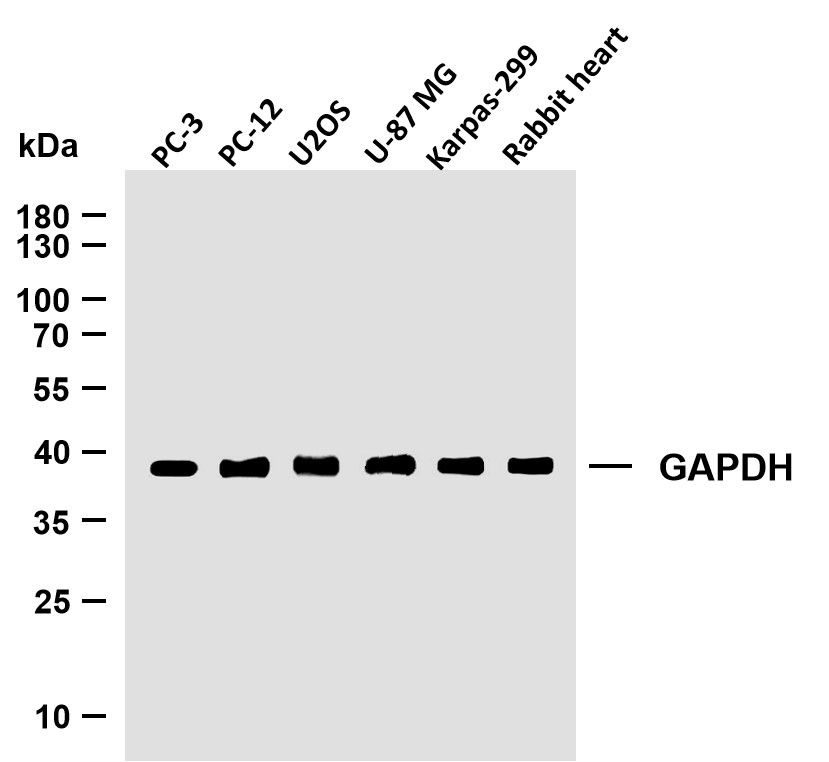
Catalog: YP0151
Size
Price
Status
Qty.
200μL
$600.00
In stock
0
100μL
$340.00
In stock
0
50μL
$190.00
In stock
0
Add to cart


Collected


Collect
Main Information
Target
IκB-α
Host Species
Rabbit
Reactivity
Human, Mouse, Rat, Monkey
Applications
WB, IHC, IF, ELISA
MW
about 40kD (Observed)
Conjugate/Modification
Phospho
Detailed Information
Recommended Dilution Ratio
WB 1:500-1:2000; IHC 1:100-1:300; IF 1:200-1:1000; ELISA 1:10000; Not yet tested in other applications.
Formulation
Liquid in PBS containing 50% glycerol, 0.5% BSA and 0.02% sodium azide.
Specificity
This antibody detects endogenous levels of IκB-α only when phosphorylated at Ser32 or Ser36.and dually phosphorylated at two sites.The name of modified sites may be influenced by many factors, such as species (the modified site was not originally found in human samples) and the change of protein sequence (the previous protein sequence is incomplete, and the protein sequence may be prolonged with the development of protein sequencing technology). When naming, we will use the "numbers" in historical reference to keep the sites consistent with the reports. The antibody binds to the following modification sequence (lowercase letters are modification sites):RHDsGLDsM
Purification
The antibody was affinity-purified from rabbit antiserum by affinity-chromatography using epitope-specific immunogen.
Storage
-15°C to -25°C/1 year(Do not lower than -25°C)
Concentration
1 mg/ml
MW(Observed)
about 40kD
Modification
Phospho
Clonality
Polyclonal
Isotype
IgG
Related Products
Antigen&Target Information
Immunogen:
The antiserum was produced against synthesized peptide derived from human IkappaB-alpha around the phosphorylation site of Ser32/Ser36. AA range:15-64
show all
Specificity:
This antibody detects endogenous levels of IκB-α only when phosphorylated at Ser32 or Ser36.and dually phosphorylated at two sites.The name of modified sites may be influenced by many factors, such as species (the modified site was not originally found in human samples) and the change of protein sequence (the previous protein sequence is incomplete, and the protein sequence may be prolonged with the development of protein sequencing technology). When naming, we will use the "numbers" in historical reference to keep the sites consistent with the reports. The antibody binds to the following modification sequence (lowercase letters are modification sites):RHDsGLDsM
show all
Gene Name:
NFKBIA IKBA MAD3 NFKBI
show all
Protein Name:
NF-kappa-B inhibitor alpha
show all
Other Name:
NFKBIA ;
IKBA ;
MAD3 ;
NFKBI ;
NF-kappa-B inhibitor alpha ;
I-kappa-B-alpha ;
IkB-alpha ;
IkappaBalpha ;
Major histocompatibility complex enhancer-binding protein MAD3
IKBA ;
MAD3 ;
NFKBI ;
NF-kappa-B inhibitor alpha ;
I-kappa-B-alpha ;
IkB-alpha ;
IkappaBalpha ;
Major histocompatibility complex enhancer-binding protein MAD3
show all
Database Link:
Background:
This gene encodes a member of the NF-kappa-B inhibitor family, which contain multiple ankrin repeat domains. The encoded protein interacts with REL dimers to inhibit NF-kappa-B/REL complexes which are involved in inflammatory responses. The encoded protein moves between the cytoplasm and the nucleus via a nuclear localization signal and CRM1-mediated nuclear export. Mutations in this gene have been found in ectodermal dysplasia anhidrotic with T-cell immunodeficiency autosomal dominant disease. [provided by RefSeq, Aug 2011],
show all
Function:
Disease:Defects in NFKBIA are the cause of ectodermal dysplasia anhidrotic with T-cell immunodeficiency autosomal dominant (ADEDAID) [MIM:612132]. Ectodermal dysplasia defines a heterogeneous group of disorders due to abnormal development of two or more ectodermal structures. ADEDAID is an ectodermal dysplasia associated with decreased production of pro-inflammatory cytokines and certain interferons, rendering patients susceptible to infection.,Function:Inhibits the activity of dimeric NF-kappa-B/REL complexes by trapping REL dimers in the cytoplasm through masking of their nuclear localization signals. On cellular stimulation by immune and proinflammatory responses, becomes phosphorylated promoting ubiquitination and degradation, enabling the dimeric RELA to tranlocate to the nucleus and activate transcription.,induction:Induced in adherent monocytes.,online information:NFKBIA mutation db,PTM:Phosphorylated; disables inhibition of NF-kappa-B DNA-binding activity.,PTM:Sumoylated; sumoylation requires the presence of the nuclear import signal.,PTM:Ubiquitinated; subsequent to stimulus-dependent phosphorylation on serines.,similarity:Belongs to the NF-kappa-B inhibitor family.,similarity:Contains 5 ANK repeats.,subcellular location:Shuttles between the nucleus and the cytoplasm by a nuclear localization signal (NLS) and a CRM1-dependent nuclear export.,subunit:Interacts with RELA; the interaction requires the nuclear import signal. Interacts with NKIRAS1 and NKIRAS2. Part of a 70-90 kDa complex at least consisting of CHUK, IKBKB, NFKBIA, RELA, IKBKAP and MAP3K14. Interacts with HBV protein X. Interacts with RWDD3; the interaction enhances sumoylation.,
show all
Cellular Localization:
Cytoplasm. Nucleus. Shuttles between the nucleus and the cytoplasm by a nuclear localization signal (NLS) and a CRM1-dependent nuclear export. .
show all
Tissue Expression:
Brain,Kidney,Lymph node,Monocyte,
show all
Research Areas:
>>cAMP signaling pathway ;
>>Chemokine signaling pathway ;
>>NF-kappa B signaling pathway ;
>>Apoptosis ;
>>Osteoclast differentiation ;
>>Toll-like receptor signaling pathway ;
>>NOD-like receptor signaling pathway ;
>>RIG-I-like receptor signaling pathway ;
>>Cytosolic DNA-sensing pathway ;
>>C-type lectin receptor signaling pathway ;
>>IL-17 signaling pathway ;
>>Th1 and Th2 cell differentiation ;
>>Th17 cell differentiation ;
>>T cell receptor signaling pathway ;
>>B cell receptor signaling pathway ;
>>TNF signaling pathway ;
>>Neurotrophin signaling pathway ;
>>Adipocytokine signaling pathway ;
>>Relaxin signaling pathway ;
>>Insulin resistance ;
>>Alcoholic liver disease ;
>>Epithelial cell signaling in Helicobacter pylori infection ;
>>Pathogenic Escherichia coli infection ;
>>Shigellosis ;
>>Salmonella infection ;
>>Legionellosis ;
>>Yersinia infection ;
>>Leishmaniasis ;
>>Chagas disease ;
>>Toxoplasmosis ;
>>Hepatitis C ;
>>Hepatitis B ;
>>Measles ;
>>Human cytomegalovirus infection ;
>>Influenza A ;
>>Human T-cell leukemia virus 1 infection ;
>>Kaposi sarcoma-associated herpesvirus infection ;
>>Herpes simplex virus 1 infection ;
>>Epstein-Barr virus infection ;
>>Human immunodeficiency virus 1 infection ;
>>Coronavirus disease - COVID-19 ;
>>Pathways in cancer ;
>>Viral carcinogenesis ;
>>Chemical carcinogenesis - reactive oxygen species ;
>>Prostate cancer ;
>>Chronic myeloid leukemia ;
>>Small cell lung cancer ;
>>PD-L1 expression and PD-1 checkpoint pathway in cancer ;
>>Lipid and atherosclerosis
>>Chemokine signaling pathway ;
>>NF-kappa B signaling pathway ;
>>Apoptosis ;
>>Osteoclast differentiation ;
>>Toll-like receptor signaling pathway ;
>>NOD-like receptor signaling pathway ;
>>RIG-I-like receptor signaling pathway ;
>>Cytosolic DNA-sensing pathway ;
>>C-type lectin receptor signaling pathway ;
>>IL-17 signaling pathway ;
>>Th1 and Th2 cell differentiation ;
>>Th17 cell differentiation ;
>>T cell receptor signaling pathway ;
>>B cell receptor signaling pathway ;
>>TNF signaling pathway ;
>>Neurotrophin signaling pathway ;
>>Adipocytokine signaling pathway ;
>>Relaxin signaling pathway ;
>>Insulin resistance ;
>>Alcoholic liver disease ;
>>Epithelial cell signaling in Helicobacter pylori infection ;
>>Pathogenic Escherichia coli infection ;
>>Shigellosis ;
>>Salmonella infection ;
>>Legionellosis ;
>>Yersinia infection ;
>>Leishmaniasis ;
>>Chagas disease ;
>>Toxoplasmosis ;
>>Hepatitis C ;
>>Hepatitis B ;
>>Measles ;
>>Human cytomegalovirus infection ;
>>Influenza A ;
>>Human T-cell leukemia virus 1 infection ;
>>Kaposi sarcoma-associated herpesvirus infection ;
>>Herpes simplex virus 1 infection ;
>>Epstein-Barr virus infection ;
>>Human immunodeficiency virus 1 infection ;
>>Coronavirus disease - COVID-19 ;
>>Pathways in cancer ;
>>Viral carcinogenesis ;
>>Chemical carcinogenesis - reactive oxygen species ;
>>Prostate cancer ;
>>Chronic myeloid leukemia ;
>>Small cell lung cancer ;
>>PD-L1 expression and PD-1 checkpoint pathway in cancer ;
>>Lipid and atherosclerosis
show all
Signaling Pathway
Cellular Processes >> Cell growth and death >> Apoptosis
Organismal Systems >> Immune system >> Toll-like receptor signaling pathway
Organismal Systems >> Immune system >> NOD-like receptor signaling pathway
Organismal Systems >> Immune system >> RIG-I-like receptor signaling pathway
Organismal Systems >> Immune system >> Cytosolic DNA-sensing pathway
Organismal Systems >> Immune system >> T cell receptor signaling pathway
Organismal Systems >> Immune system >> Th1 and Th2 cell differentiation
Organismal Systems >> Immune system >> Th17 cell differentiation
Organismal Systems >> Immune system >> IL-17 signaling pathway
Organismal Systems >> Immune system >> B cell receptor signaling pathway
Organismal Systems >> Immune system >> Chemokine signaling pathway
Organismal Systems >> Endocrine system >> Adipocytokine signaling pathway
Organismal Systems >> Endocrine system >> Relaxin signaling pathway
Organismal Systems >> Nervous system >> Neurotrophin signaling pathway
Organismal Systems >> Development and regeneration >> Osteoclast differentiation
Human Diseases >> Cancer: overview >> Pathways in cancer
Human Diseases >> Cancer: overview >> PD-L1 expression and PD-1 checkpoint pathway in cancer
Human Diseases >> Cancer: specific types >> Chronic myeloid leukemia
Human Diseases >> Cancer: specific types >> Prostate cancer
Human Diseases >> Cancer: specific types >> Small cell lung cancer
Environmental Information Processing >> Signal transduction >> NF-kappa B signaling pathway
Environmental Information Processing >> Signal transduction >> TNF signaling pathway
Environmental Information Processing >> Signal transduction >> cAMP signaling pathway
Reference Citation({{totalcount}})
Catalog: YP0151
Size
Price
Status
Qty.
200μL
$600.00
In stock
0
100μL
$340.00
In stock
0
50μL
$190.00
In stock
0
Add to cart


Collected


Collect
Recently Viewed Products
Clear allPRODUCTS
CUSTOMIZED
ABOUT US
Toggle night Mode
{{pinfoXq.title || ''}}
Catalog: {{pinfoXq.catalog || ''}}
Filter:
All
{{item.name}}
{{pinfo.title}}
-{{pinfo.catalog}}
Main Information
Target
{{pinfo.target}}
Reactivity
{{pinfo.react}}
Applications
{{pinfo.applicat}}
Conjugate/Modification
{{pinfo.coupling}}/{{pinfo.modific}}
MW (kDa)
{{pinfo.mwcalc}}
Host Species
{{pinfo.hostspec}}
Isotype
{{pinfo.isotype}}
Product {{index}}/{{pcount}}
Prev
Next
{{pvTitle}}
Scroll wheel zooms the picture
{{pvDescr}}