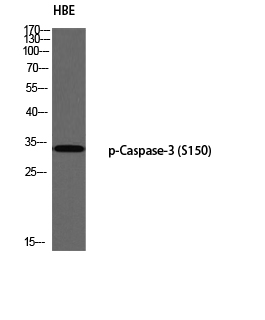
Catalog: KA1157C
Size
Price
Status
Qty.
96well
$470.00
In stock
0
Add to cart


Collected


Collect
Main Information
Reactivity
Human, Mouse, Rat
Applications
ELISA
Conjugate/Modification
Phospho
Detailed Information
Storage
2-8°C/6 months,Ship by ice bag
Modification
Phospho
Detection Method
Colorimetric
Related Products
Antigen&Target Information
Gene Name:
CASP3
show all
Other Name:
Caspase-3 ;
CASP-3 ;
Apopain ;
Cysteine protease CPP32 ;
CPP-32 ;
Protein Yama ;
SREBP cleavage activity 1 ;
SCA-1 ;
[Cleaved into: Caspase-3 subunit p17 ;
Caspase-3 subunit p12]
CASP-3 ;
Apopain ;
Cysteine protease CPP32 ;
CPP-32 ;
Protein Yama ;
SREBP cleavage activity 1 ;
SCA-1 ;
[Cleaved into: Caspase-3 subunit p17 ;
Caspase-3 subunit p12]
show all
Database Link:
Background:
catalytic activity:Strict requirement for an Asp residue at positions P1 and P4. It has a preferred cleavage sequence of Asp-Xaa-Xaa-Asp-|- with a hydrophobic amino-acid residue at P2 and a hydrophilic amino-acid residue at P3, although Val or Ala are also accepted at this position.,enzyme regulation:Inhibited by isatin sulfonamides.,function:Involved in the activation cascade of caspases responsible for apoptosis execution. At the onset of apoptosis it proteolytically cleaves poly(ADP-ribose) polymerase (PARP) at a '216-Asp-|-Gly-217' bond. Cleaves and activates sterol regulatory element binding proteins (SREBPs) between the basic helix-loop-helix leucine zipper domain and the membrane attachment domain. Cleaves and activates caspase-6, -7 and -9. Involved in the cleavage of huntingtin.,PTM:Cleavage by granzyme B, caspase-6, caspase-8 and caspase-10 generates the two active subunits. Additional processing of the propeptides is likely due to the autocatalytic activity of the activated protease. Active heterodimers between the small subunit of caspase-7 protease and the large subunit of caspase-3 also occur and vice versa.,PTM:S-nitrosylated on its catalytic site cysteine in unstimulated human cell lines and denitrosylated upon activation of the Fas apoptotic pathway, associated with an increase in intracellular caspase activity. Fas therefore activates caspase-3 not only by inducing the cleavage of the caspase zymogen to its active subunits, but also by stimulating the denitrosylation of its active site thiol.,similarity:Belongs to the peptidase C14A family.,subunit:Heterotetramer that consists of two anti-parallel arranged heterodimers, each one formed by a 17 kDa (p17) and a 12 kDa (p12) subunit.,tissue specificity:Highly expressed in lung, spleen, heart, liver and kidney. Moderate levels in brain and skeletal muscle, and low in testis. Also found in many cell lines, highest expression in cells of the immune system.,
show all
Function:
regulation of cyclin-dependent protein kinase activity, DNA catabolic process, endonucleolytic, leukocyte homeostasis,B cell homeostasis, release of cytochrome c from mitochondria, lymphocyte homeostasis, negative regulation of immune system process, regulation of leukocyte activation, negative regulation of leukocyte activation, DNA metabolic process, DNA catabolic process, DNA fragmentation involved in apoptosis, negative regulation of protein kinase activity, proteolysis, apoptosis, induction of apoptosis, cell structure disassembly during apoptosis, response to DNA damage stimulus, nucleus organization, mitochondrion organization, ectoderm development, heart development,sensory perception, sensory perception of sound, cell death, negative regulation of cell proliferation, epidermis development, induction of apoptosis by extracellular signals, induction of apoptosis by intracellular signals, induction of apoptosis by oxidative stress, apoptotic mitochondrial changes, macromolecule catabolic process, response to radiation, response to UV, response to light stimulus, response to wounding, response to abiotic stimulus, epidermal cell differentiation, response to organic substance, regulation of cell death, positive regulation of cell death,programmed cell death, induction of programmed cell death, death, regulation of phosphate metabolic process,cellular component disassembly, keratinocyte differentiation, apoptotic nuclear changes, nuclear fragmentation during apoptosis, epithelial cell differentiation, regulation of B cell proliferation, negative regulation of B cell proliferation,regulation of mononuclear cell proliferation, negative regulation of mononuclear cell proliferation, cellular response to stress, negative regulation of kinase activity, response to cytokine stimulus, response to tumor necrosis factor,regulation of cell proliferation, regulation of T cell proliferation, negative regulation of T cell proliferation, regulation of phosphorylation, homeostatic process, regulation of apoptosis, T cell homeostasis, positive regulation of apoptosis,negative regulation of apoptosis, regulation of programmed cell death, positive regulation of programmed cell death,negative regulation of programmed cell death, negative regulation of catalytic activity, regulation of kinase activity,negative regulation of molecular function, cellular macromolecule catabolic process, cell fate commitment, negative regulation of cyclin-dependent protein kinase activity, negative regulation of cell cycle, regulation of protein kinase activity, regulation of activated T cell proliferation, negative regulation of activated T cell proliferation, homeostasis of number of cells, regulation of lymphocyte proliferation, negative regulation of lymphocyte proliferation, regulation of T cell activation, regulation of B cell activation, regulation of cell activation, negative regulation of cell activation,negative regulation of T cell activation, negative regulation of B cell activation, neurological system process, cognition,sensory perception of mechanical stimulus, regulation of phosphorus metabolic process, regulation of lymphocyte activation, negative regulation of lymphocyte activation, regulation of transferase activity, negative regulation of transferase activity, neuron apoptosis, regulation of cell cycle, epithelium development, negative regulation of cell death, regulation of leukocyte proliferation, negative regulation of leukocyte proliferation,
show all
Cellular Localization:
Cytoplasm.
show all
Signaling Pathway
Cellular Processes >> Cell growth and death >> Apoptosis
Cellular Processes >> Cell growth and death >> Apoptosis - multiple species
Cellular Processes >> Cell growth and death >> p53 signaling pathway
Organismal Systems >> Immune system >> Cytosolic DNA-sensing pathway
Organismal Systems >> Immune system >> Natural killer cell mediated cytotoxicity
Organismal Systems >> Immune system >> IL-17 signaling pathway
Human Diseases >> Cancer: overview >> Pathways in cancer
Human Diseases >> Cancer: overview >> MicroRNAs in cancer
Human Diseases >> Cancer: specific types >> Colorectal cancer
Human Diseases >> Cancer: specific types >> Small cell lung cancer
Human Diseases >> Neurodegenerative disease >> Alzheimer disease
Human Diseases >> Neurodegenerative disease >> Parkinson disease
Human Diseases >> Neurodegenerative disease >> Amyotrophic lateral sclerosis
Human Diseases >> Neurodegenerative disease >> Huntington disease
Human Diseases >> Neurodegenerative disease >> Prion disease
Human Diseases >> Neurodegenerative disease >> Pathways of neurodegeneration - multiple diseases
Environmental Information Processing >> Signal transduction >> MAPK signaling pathway
Environmental Information Processing >> Signal transduction >> TNF signaling pathway
Reference Citation({{totalcount}})
Catalog: KA1157C
Size
Price
Status
Qty.
96well
$470.00
In stock
0
Add to cart


Collected


Collect
Recently Viewed Products
Clear allPRODUCTS
CUSTOMIZED
ABOUT US
Toggle night Mode
{{pinfoXq.title || ''}}
Catalog: {{pinfoXq.catalog || ''}}
Filter:
All
{{item.name}}
{{pinfo.title}}
-{{pinfo.catalog}}
Main Information
Target
{{pinfo.target}}
Reactivity
{{pinfo.react}}
Applications
{{pinfo.applicat}}
Conjugate/Modification
{{pinfo.coupling}}/{{pinfo.modific}}
MW (kDa)
{{pinfo.mwcalc}}
Host Species
{{pinfo.hostspec}}
Isotype
{{pinfo.isotype}}
Product {{index}}/{{pcount}}
Prev
Next
{{pvTitle}}
Scroll wheel zooms the picture
{{pvDescr}}