Catalog: YN7682
Size
Price
Status
Qty.
200μL
$450.00
In stock
0
100μL
$280.00
In stock
0
40μL
$160.00
In stock
0
Add to cart


Collected


Collect
Main Information
Target
ADPRS
Host Species
Rabbit
Reactivity
Human, Mouse
Applications
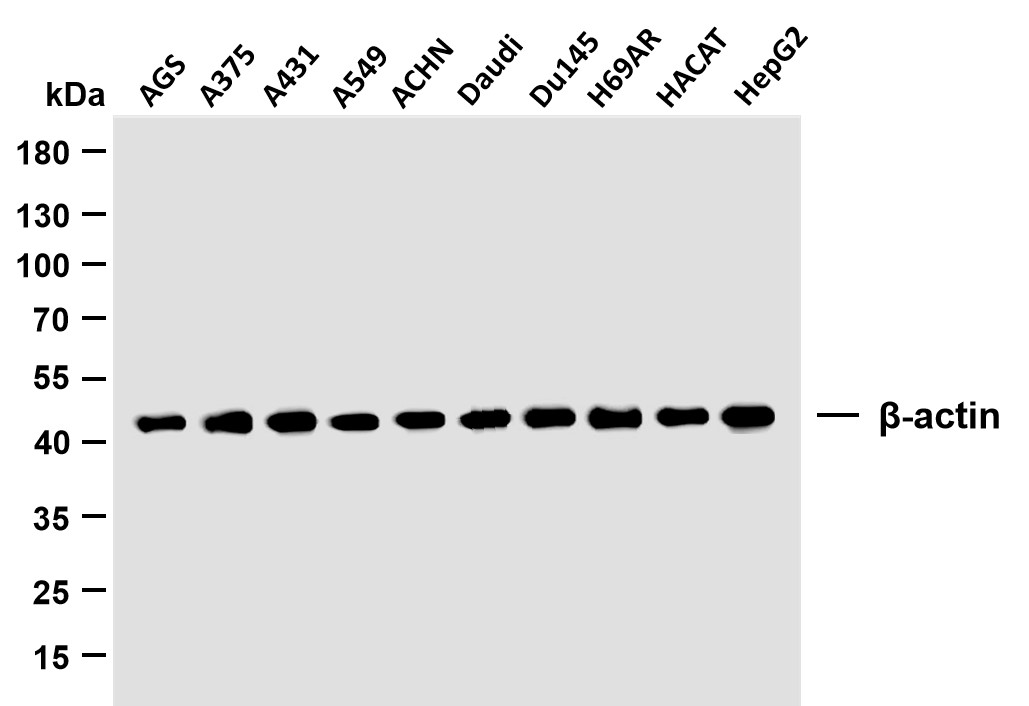
WB
MW
40kD (Calculated)
Conjugate/Modification
Unmodified
Detailed Information
Recommended Dilution Ratio
WB 1:500-2000
Formulation
Liquid in PBS containing 50% glycerol, 0.5% BSA and 0.02% sodium azide.
Specificity
This antibody detects endogenous levels of ARH3 at Human, Mouse
Purification
The antibody was affinity-purified from rabbit antiserum by affinity-chromatography using epitope-specific immunogen.
Storage
-15°C to -25°C/1 year(Do not lower than -25°C)
Concentration
1 mg/ml
MW(Calculated)
40kD
Modification
Unmodified
Clonality
Polyclonal
Isotype
IgG
Related Products
Antigen&Target Information
Immunogen:
Synthesized peptide derived from human ARH3
show all
Specificity:
This antibody detects endogenous levels of ARH3 at Human, Mouse
show all
Gene Name:
ADPRHL2 ARH3
show all
Protein Name:
Poly(ADP-ribose) glycohydrolase ARH3 (ADP-ribosylhydrolase 3) ([Protein ADP-ribosylarginine] hydrolase-like protein 2)
show all
Function:
ADP-ribosylhydrolase that preferentially hydrolyzes the scissile alpha-O-linkage attached to the anomeric C1'' position of ADP-ribose and acts on different substrates, such as proteins ADP-ribosylated on serine and threonine, free poly(ADP-ribose) and O-acetyl-ADP-D-ribose . Specifically acts as a serine mono-ADP-ribosylhydrolase by mediating the removal of mono-ADP-ribose attached to serine residues on proteins, thereby playing a key role in DNA damage response . Serine ADP-ribosylation of proteins constitutes the primary form of ADP-ribosylation of proteins in response to DNA damage . Does not hydrolyze ADP-ribosyl-arginine, -cysteine, -diphthamide, or -asparagine bonds . Also able to degrade protein free poly(ADP-ribose), which is synthesized in response to DNA damage: free poly(ADP-ribose) acts as a potent cell death signal and its degradation by ADPRHL2 protects cells from poly(ADP-ribose)-dependent cell death, a process named parthanatos . Also hydrolyzes free poly(ADP-ribose) in mitochondria . Specifically digests O-acetyl-ADP-D-ribose, a product of deacetylation reactions catalyzed by sirtuins . Specifically degrades 1''-O-acetyl-ADP-D-ribose isomer, rather than 2''-O-acetyl-ADP-D-ribose or 3''-O-acetyl-ADP-D-ribose isomers .
show all
Cellular Localization:
Nucleus . Cytoplasm . Chromosome . Mitochondrion matrix . Recruited to DNA lesion regions following DNA damage; ADP-D-ribose-recognition is required for recruitment to DNA damage sites. .
show all
Tissue Expression:
Ubiquitous (PubMed:16278211). Expressed in skin fibroblasts (PubMed:30830864).
show all
Reference Citation({{totalcount}})
Catalog: YN7682
Size
Price
Status
Qty.
200μL
$450.00
In stock
0
100μL
$280.00
In stock
0
40μL
$160.00
In stock
0
Add to cart


Collected


Collect
Recently Viewed Products
Clear allPRODUCTS
CUSTOMIZED
ABOUT US
Toggle night Mode
{{pinfoXq.title || ''}}
Catalog: {{pinfoXq.catalog || ''}}
Filter:
All
{{item.name}}
{{pinfo.title}}
-{{pinfo.catalog}}
Main Information
Target
{{pinfo.target}}
Reactivity
{{pinfo.react}}
Applications
{{pinfo.applicat}}
Conjugate/Modification
{{pinfo.coupling}}/{{pinfo.modific}}
MW (kDa)
{{pinfo.mwcalc}}
Host Species
{{pinfo.hostspec}}
Isotype
{{pinfo.isotype}}
Product {{index}}/{{pcount}}
Prev
Next
{{pvTitle}}
Scroll wheel zooms the picture
{{pvDescr}}