Catalog: YK0071
Size
Price
Status
Qty.
200μL
$600.00
In stock
0
100μL
$340.00
In stock
0
50μL
$190.00
In stock
0
Add to cart


Collected


Collect
Main Information
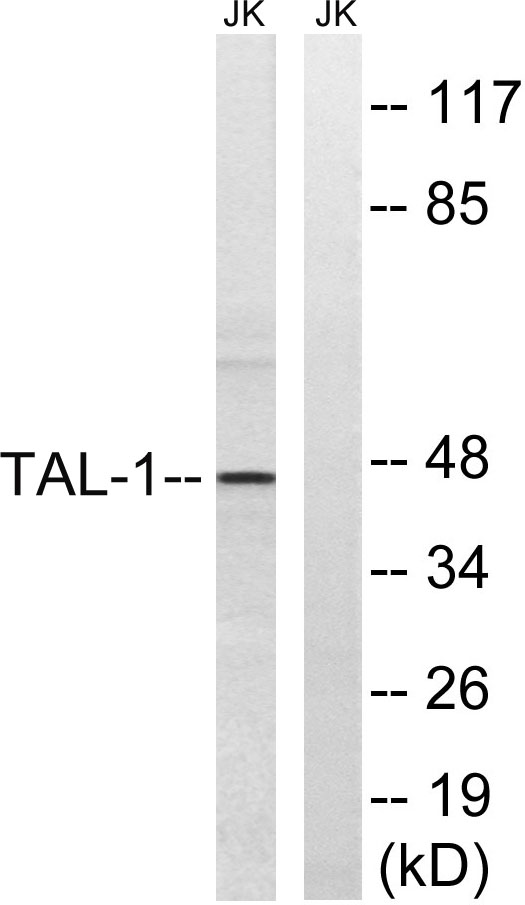
Target
TAL1/2
Host Species
Rabbit
Reactivity
Human, Mouse, Rat
Applications
WB, ELISA
MW
45kD (Observed)
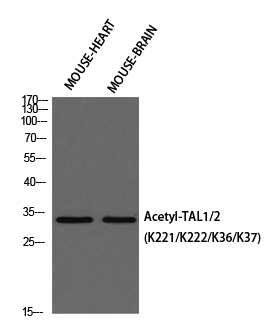
Conjugate/Modification
Acetyl
Detailed Information
Recommended Dilution Ratio
WB 1:500-1:2000; ELISA 1:10000; Not yet tested in other applications.
Formulation
Liquid in PBS containing 50% glycerol, 0.5% BSA and 0.02% sodium azide.
Specificity
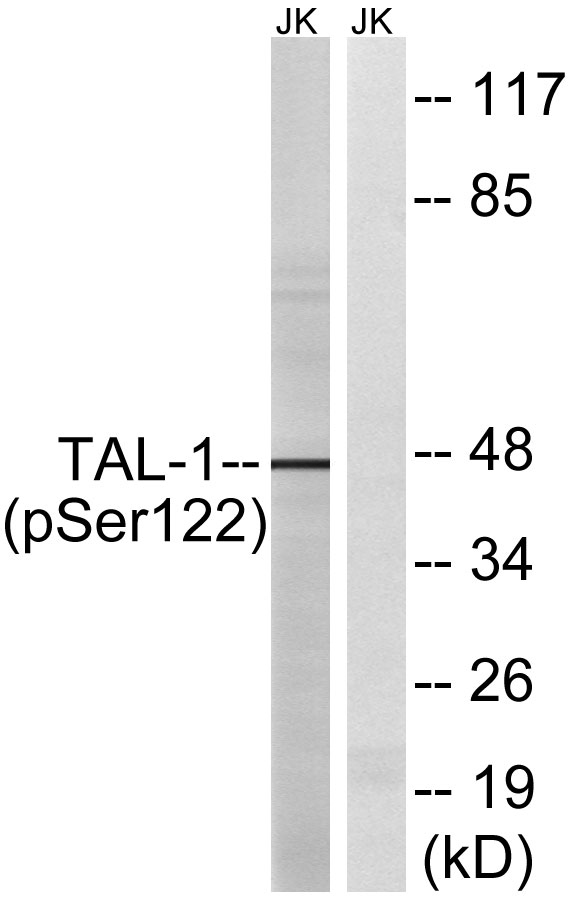
This antibody detects endogenous levels of TAL1 only when phosphorylated at Human:K221+K222, Mouse:K221+K222, Rat:K221+K222,and dually phosphorylated at two sites. It can also detects endogenous levels of TAL2 only when phosphorylated at Human:K36+K37, Mouse:K36+K37, Rat:K36+K37,and dually phosphorylated at two sites.The name of modified sites may be influenced by many factors, such as species (the modified site was not originally found in human samples) and the change of protein sequence (the previous protein sequence is incomplete, and the protein sequence may be prolonged with the development of protein sequencing technology). When naming, we will use the "numbers" in historical reference to keep the sites consistent with the reports. The antibody binds to the following modification sequence (lowercase letters are modification sites):DkkLS
Purification
The antibody was affinity-purified from rabbit antiserum by affinity-chromatography using epitope-specific immunogen.
Storage
-15°C to -25°C/1 year(Do not lower than -25°C)
Concentration
1 mg/ml
MW(Observed)
45kD
Modification
Acetyl
Clonality
Polyclonal
Isotype
IgG
Related Products
Antigen&Target Information
Immunogen:
Synthesized acetyl-peptide derived from human TAL1/2 around the acetylation site of K221.
show all
Specificity:
This antibody detects endogenous levels of TAL1 only when phosphorylated at Human:K221+K222, Mouse:K221+K222, Rat:K221+K222,and dually phosphorylated at two sites. It can also detects endogenous levels of TAL2 only when phosphorylated at Human:K36+K37, Mouse:K36+K37, Rat:K36+K37,and dually phosphorylated at two sites.The name of modified sites may be influenced by many factors, such as species (the modified site was not originally found in human samples) and the change of protein sequence (the previous protein sequence is incomplete, and the protein sequence may be prolonged with the development of protein sequencing technology). When naming, we will use the "numbers" in historical reference to keep the sites consistent with the reports. The antibody binds to the following modification sequence (lowercase letters are modification sites):DkkLS
show all
Gene Name:
TAL1/TAL2
show all
Protein Name:
T-cell acute lymphocytic leukemia protein 1 homolog/T-cell acute lymphocytic leukemia protein 2
show all
Other Name:
TAL1 ;
BHLHA17 ;
SCL ;
TCL5 ;
T-cell acute lymphocytic leukemia protein 1 ;
TAL-1 ;
Class A basic helix-loop-helix protein 17 ;
bHLHa17 ;
Stem cell protein ;
T-cell leukemia/lymphoma protein 5 ;
TAL2 ;
BHLHA19 ;
T-cell acute lymphocytic leukemia protein 2 ;
TAL-2 ;
Class A basic helix-loop-helix protein 19 ;
bHLHa19
BHLHA17 ;
SCL ;
TCL5 ;
T-cell acute lymphocytic leukemia protein 1 ;
TAL-1 ;
Class A basic helix-loop-helix protein 17 ;
bHLHa17 ;
Stem cell protein ;
T-cell leukemia/lymphoma protein 5 ;
TAL2 ;
BHLHA19 ;
T-cell acute lymphocytic leukemia protein 2 ;
TAL-2 ;
Class A basic helix-loop-helix protein 19 ;
bHLHa19
show all
Background:
alternative products:The splicing pattern is cell-lineage dependent,disease:A chromosomal aberration involving TAL1 may be a cause of some T-cell acute lymphoblastic leukemias (T-ALL). Translocation t(1;14)(p32;q11) with T-cell receptor alpha chain (TCRA) genes.,domain:The helix-loop-helix domain is necessary and sufficient for the interaction with DRG1.,function:Implicated in the genesis of hemopoietic malignancies. It may play an important role in hemopoietic differentiation. Serves as a positive regulator of erythroid differentiation.,PTM:Phosphorylated on serine residues. Phosphorylation of Ser-122 is strongly stimulated by hypoxia.,PTM:Ubiquitinated; subsequent to hypoxia-dependent phosphorylation of Ser-122, ubiquitination targets the protein for rapid degradation via the ubiquitin system. This process may be characteristic for microvascular endothelial cells, since it could not be observed in large vessel endothelial cells.,similarity:Contains 1 basic helix-loop-helix (bHLH) domain.,subunit:Efficient DNA binding requires dimerization with another bHLH protein. Forms heterodimers with TCF3. Binds to the LIM domain containing protein LMO2 and to DRG1. Can assemble in a complex with LDB1 and LMO2. Component of a TAL-1 complex composed at least of CBFA2T3, LDB1, TAL1 and TCF3.,tissue specificity:Leukemic stem cell.,
show all
Function:
Alternative products:The splicing pattern is cell-lineage dependent,Disease:A chromosomal aberration involving TAL1 may be a cause of some T-cell acute lymphoblastic leukemias (T-ALL). Translocation t(1;14)(p32;q11) with T-cell receptor alpha chain (TCRA) genes.,Domain:The helix-loop-helix domain is necessary and sufficient for the interaction with DRG1.,Function:Implicated in the genesis of hemopoietic malignancies. It may play an important role in hemopoietic differentiation. Serves as a positive regulator of erythroid differentiation.,PTM:Phosphorylated on serine residues. Phosphorylation of Ser-122 is strongly stimulated by hypoxia.,PTM:Ubiquitinated; subsequent to hypoxia-dependent phosphorylation of Ser-122, ubiquitination targets the protein for rapid degradation via the ubiquitin system. This process may be characteristic for microvascular endothelial cells, since it could not be observed in large vessel endothelial cells.,similarity:Contains 1 basic helix-loop-helix (bHLH) domain.,subunit:Efficient DNA binding requires dimerization with another bHLH protein. Forms heterodimers with TCF3. Binds to the LIM domain containing protein LMO2 and to DRG1. Can assemble in a complex with LDB1 and LMO2. Component of a TAL-1 complex composed at least of CBFA2T3, LDB1, TAL1 and TCF3.,tissue specificity:Leukemic stem cell.,
show all
Cellular Localization:
Nucleus .
show all
Tissue Expression:
show all
Reference Citation({{totalcount}})
Catalog: YK0071
Size
Price
Status
Qty.
200μL
$600.00
In stock
0
100μL
$340.00
In stock
0
50μL
$190.00
In stock
0
Add to cart


Collected


Collect
Recently Viewed Products
Clear allPRODUCTS
CUSTOMIZED
ABOUT US
Toggle night Mode
{{pinfoXq.title || ''}}
Catalog: {{pinfoXq.catalog || ''}}
Filter:
All
{{item.name}}
{{pinfo.title}}
-{{pinfo.catalog}}
Main Information
Target
{{pinfo.target}}
Reactivity
{{pinfo.react}}
Applications
{{pinfo.applicat}}
Conjugate/Modification
{{pinfo.coupling}}/{{pinfo.modific}}
MW (kDa)
{{pinfo.mwcalc}}
Host Species
{{pinfo.hostspec}}
Isotype
{{pinfo.isotype}}
Product {{index}}/{{pcount}}
Prev
Next
{{pvTitle}}
Scroll wheel zooms the picture
{{pvDescr}}