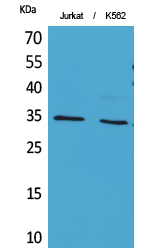
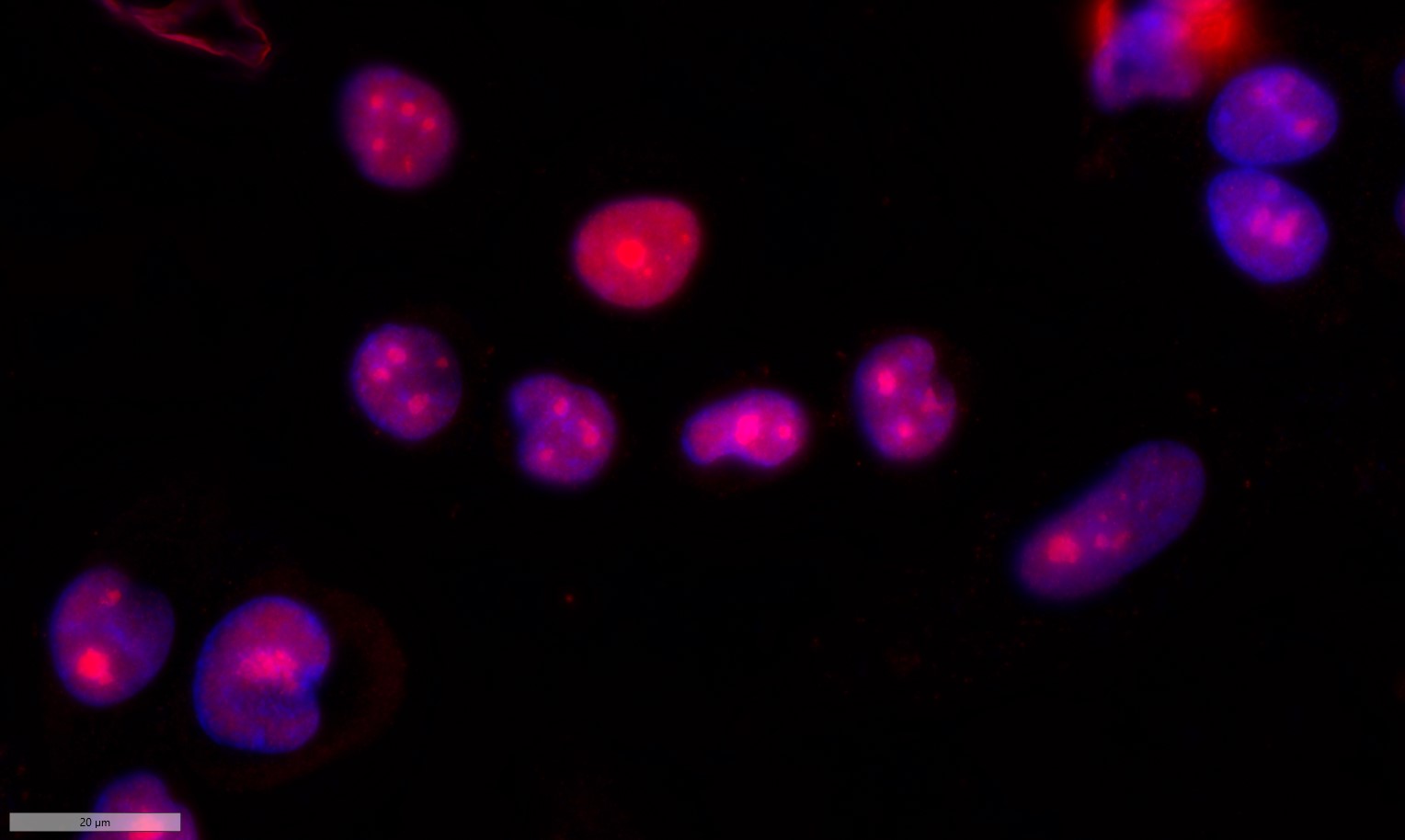
Catalog: KA3575C
Size
Price
Status
Qty.
96well
$330.00
In stock
0
Add to cart


Collected


Collect
Main Information
Reactivity
Human, Mouse, Rat
Applications
ELISA
Conjugate/Modification
Unmodified
Detailed Information
Storage
2-8°C/6 months,Ship by ice bag
Modification
Unmodified
Detection Method
Colorimetric
Related Products
Antigen&Target Information
Gene Name:
APEX1
show all
Other Name:
DNA- ;
apurinic or apyrimidinic site ;
lyase ;
EC 4.2.99.18 ;
APEX nuclease ;
APEN ;
Apurinic-apyrimidinic endonuclease 1 ;
AP endonuclease 1 ;
APE-1 ;
REF-1 ;
Redox factor-1 ;
[Cleaved into: DNA- ;
apurinic or apyrimidinic site ;
lyase, mitochondrial]
apurinic or apyrimidinic site ;
lyase ;
EC 4.2.99.18 ;
APEX nuclease ;
APEN ;
Apurinic-apyrimidinic endonuclease 1 ;
AP endonuclease 1 ;
APE-1 ;
REF-1 ;
Redox factor-1 ;
[Cleaved into: DNA- ;
apurinic or apyrimidinic site ;
lyase, mitochondrial]
show all
Background:
catalytic activity:The C-O-P bond 3' to the apurinic or apyrimidinic site in DNA is broken by a beta-elimination reaction, leaving a 3'-terminal unsaturated sugar and a product with a terminal 5'-phosphate.,function:Repairs oxidative DNA damages in vitro. May have a role in protection against cell lethality and suppression of mutations. Removes the blocking groups from the 3'-termini of the DNA strand breaks generated by ionizing radiations and bleomycin.,similarity:Belongs to the DNA repair enzymes AP/exoA family.,subunit:Monomer. Component of the SET complex, which also contains SET, ANP32A, HMGB2 and NME1.,
show all
Function:
DNA metabolic process, DNA repair, base-excision repair, transcription, transcription, DNA-dependent, transcription from RNA polymerase II promoter, response to DNA damage stimulus, cellular homeostasis, RNA biosynthetic process,cellular response to stress, homeostatic process, cell redox homeostasis, regulation of binding, regulation of DNA binding,
show all
Cellular Localization:
Nucleus. Nucleus, nucleolus. Nucleus speckle. Endoplasmic reticulum. Cytoplasm. Detected in the cytoplasm of B-cells stimulated to switch (By similarity). Colocalized with SIRT1 in the nucleus. Colocalized with YBX1 in nuclear speckles after genotoxic stress. Together with OGG1 is recruited to nuclear speckles in UVA-irradiated cells. Colocalized with nucleolin and NPM1 in the nucleolus. Its nucleolar localization is cell cycle dependent and requires active rRNA transcription. Colocalized with calreticulin in the endoplasmic reticulum. Translocation from the nucleus to the cytoplasm is stimulated in presence of nitric oxide (NO) and function in a CRM1-dependent manner, possibly as a consequence of demasking a nuclear export signal (amino acid position 64-80). S-nitrosylation at Cys-93 and Cys-310 regulates its nuclear-cytosolic shuttling. Ubiquitinated form is localized predominantly in the cytoplasm. .; [DNA-(apurinic or apyrimidinic site) endonuclease, mitochondrial]: Mitochondrion. The cleaved APEX2 is only detected in mitochondria (By similarity). Translocation from the cytoplasm to the mitochondria is mediated by ROS signaling and cleavage mediated by granzyme A. Tom20-dependent translocated mitochondrial APEX1 level is significantly increased after genotoxic stress. .
show all
Reference Citation({{totalcount}})
Catalog: KA3575C
Size
Price
Status
Qty.
96well
$330.00
In stock
0
Add to cart


Collected


Collect
Recently Viewed Products
Clear allPRODUCTS
CUSTOMIZED
ABOUT US
Toggle night Mode
{{pinfoXq.title || ''}}
Catalog: {{pinfoXq.catalog || ''}}
Filter:
All
{{item.name}}
{{pinfo.title}}
-{{pinfo.catalog}}
Main Information
Target
{{pinfo.target}}
Reactivity
{{pinfo.react}}
Applications
{{pinfo.applicat}}
Conjugate/Modification
{{pinfo.coupling}}/{{pinfo.modific}}
MW (kDa)
{{pinfo.mwcalc}}
Host Species
{{pinfo.hostspec}}
Isotype
{{pinfo.isotype}}
Product {{index}}/{{pcount}}
Prev
Next
{{pvTitle}}
Scroll wheel zooms the picture
{{pvDescr}}