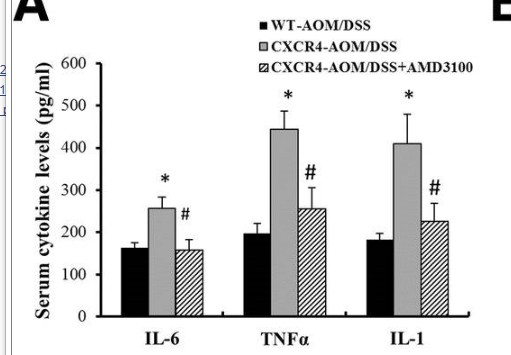
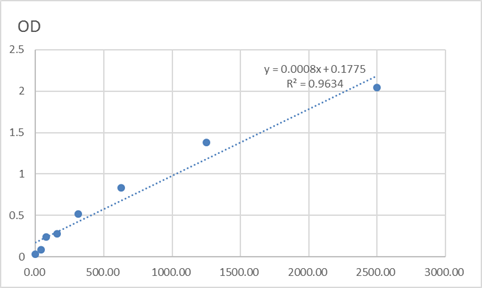
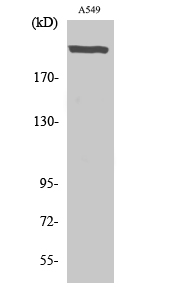Catalog: KA3404C
Size
Price
Status
Qty.
96well
$330.00
In stock
0
Add to cart


Collected


Collect
Main Information
Reactivity
Human, Mouse
Applications
ELISA
Conjugate/Modification
Unmodified
Detailed Information
Storage
2-8°C/6 months,Ship by ice bag
Modification
Unmodified
Detection Method
Colorimetric
Related Products
Antigen&Target Information
Gene Name:
UBR5
show all
Other Name:
UBR5 ;
EDD ;
EDD1 ;
HYD ;
KIAA0896 ;
E3 ubiquitin-protein ligase UBR5 ;
E3 ubiquitin-protein ligase ;
HECT domain-containing 1 ;
Hyperplastic discs protein homolog ;
hHYD ;
Progestin-induced protein
EDD ;
EDD1 ;
HYD ;
KIAA0896 ;
E3 ubiquitin-protein ligase UBR5 ;
E3 ubiquitin-protein ligase ;
HECT domain-containing 1 ;
Hyperplastic discs protein homolog ;
hHYD ;
Progestin-induced protein
show all
Background:
function:E3 ubiquitin-protein ligase which is a component of the N-end rule pathway. Recognizes and binds to proteins bearing specific amino-terminal residues that are destabilizing according to the N-end rule, leading to their ubiquitination and subsequent degradation (By similarity). May be involved in maturation and/or transcriptional regulation of mRNA. May play a role in control of cell cycle progression. May have tumor suppressor function. Regulates DNA topoisomerase II binding protein (TopBP1) in the DNA damage response. Plays an essential role in extraembryonic development.,miscellaneous:A cysteine residue is required for ubiquitin-thioester formation.,pathway:Protein modification; protein ubiquitination.,PTM:Phosphorylated upon DNA damage, probably by ATM or ATR.,similarity:Contains 1 HECT (E6AP-type E3 ubiquitin-protein ligase) domain.,similarity:Contains 1 PABC domain.,similarity:Contains 1 UBR-type zinc finger.,subunit:Binds TOPBP1.,tissue specificity:Widely expressed. Most abundant in testis and expressed at high levels in brain, pituitary and kidney.,
show all
Function:
proteolysis, ubiquitin-dependent protein catabolic process, response to DNA damage stimulus, intracellular signaling cascade, cell proliferation, macromolecule catabolic process, modification-dependent protein catabolic process, protein catabolic process, steroid hormone receptor signaling pathway, intracellular receptor-mediated signaling pathway,cellular response to stress, modification-dependent macromolecule catabolic process, cellular protein catabolic process, cellular macromolecule catabolic process, progesterone receptor signaling pathway, proteolysis involved in cellular protein catabolic process,
show all
Cellular Localization:
Nucleus.
show all
Tissue Expression:
Signaling Pathway
Reference Citation({{totalcount}})
Catalog: KA3404C
Size
Price
Status
Qty.
96well
$330.00
In stock
0
Add to cart


Collected


Collect
Recently Viewed Products
Clear allPRODUCTS
CUSTOMIZED
ABOUT US
Toggle night Mode
{{pinfoXq.title || ''}}
Catalog: {{pinfoXq.catalog || ''}}
Filter:
All
{{item.name}}
{{pinfo.title}}
-{{pinfo.catalog}}
Main Information
Target
{{pinfo.target}}
Reactivity
{{pinfo.react}}
Applications
{{pinfo.applicat}}
Conjugate/Modification
{{pinfo.coupling}}/{{pinfo.modific}}
MW (kDa)
{{pinfo.mwcalc}}
Host Species
{{pinfo.hostspec}}
Isotype
{{pinfo.isotype}}
Product {{index}}/{{pcount}}
Prev
Next
{{pvTitle}}
Scroll wheel zooms the picture
{{pvDescr}}