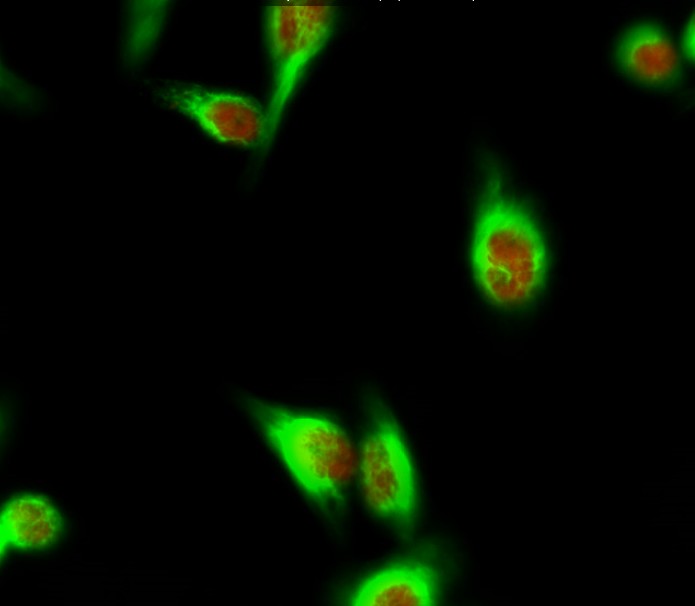
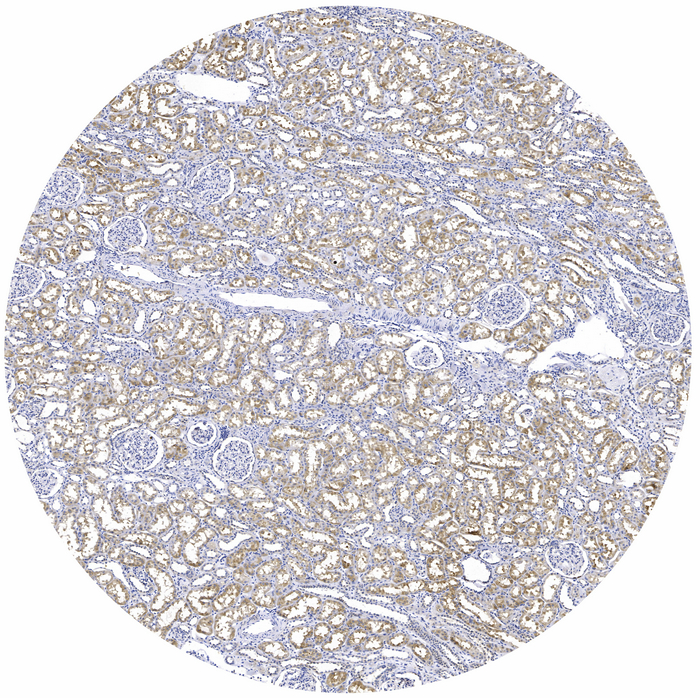
Catalog: KA3290C
Size
Price
Status
Qty.
96well
$330.00
In stock
0
Add to cart


Collected


Collect
Main Information
Reactivity
Human, Mouse
Applications
ELISA
Conjugate/Modification
Unmodified
Detailed Information
Storage
2-8°C/6 months,Ship by ice bag
Modification
Unmodified
Detection Method
Colorimetric
Related Products
Antigen&Target Information
Gene Name:
HDAC6
show all
Other Name:
Histone deacetylase 6 ;
HD6 ;
HD6 ;
show all
Background:
catalytic activity:Hydrolysis of an N(6)-acetyl-lysine residue of a histone to yield a deacetylated histone.,function:Responsible for the deacetylation of lysine residues on the N-terminal part of the core histones (H2A, H2B, H3 and H4). Histone deacetylation gives a tag for epigenetic repression and plays an important role in transcriptional regulation, cell cycle progression and developmental events. Histone deacetylases act via the formation of large multiprotein complexes (By similarity). Plays a central role in microtubule-dependent cell motility via deacetylation of tubulin.,PTM:Sumoylated in vitro.,PTM:Ubiquitinated. Its polyubiquitination however does not lead to its degradation.,similarity:Belongs to the histone deacetylase family. Type 2 subfamily.,similarity:Contains 1 UBP-type zinc finger.,subcellular location:It is mainly cytoplasmic, where it is associated with microtubules.,subunit:Interacts with CBFA2T3, HDAC11 and SIRT2. Interacts with F-actin. Interacts with BBIP10.,
show all
Function:
protein polyubiquitination, response to reactive oxygen species, chromatin organization, transcription, regulation of transcription, DNA-dependent, protein amino acid deacetylation, proteolysis, misfolded or incompletely synthesized protein catabolic process, intracellular protein transport, autophagy, response to oxidative stress, negative regulation of microtubule depolymerization, protein localization, macromolecule catabolic process, cellular response to starvation,response to toxin, response to endogenous stimulus, positive regulation of biosynthetic process, positive regulation of signal transduction, response to extracellular stimulus, response to organic substance, response to inorganic substance, regulation of hydrogen peroxide metabolic process, regulation of receptor activity, positive regulation of macromolecule biosynthetic process, positive regulation of macromolecule metabolic process, negative regulation of macromolecule metabolic process, regulation of epithelial cell migration, positive regulation of epithelial cell migration,negative regulation of organelle organization, positive regulation of cell communication, negative regulation of oxygen and reactive oxygen species metabolic process, negative regulation of hydrogen peroxide metabolic process,regulation of receptor biosynthetic process, positive regulation of receptor biosynthetic process, regulation of cell death, positive regulation of cell death, protein transport, macroautophagy, protein ubiquitination, chromatin modification, covalent chromatin modification, histone modification, histone deacetylation, peptidyl-lysine modification,regulation of proteolysis, protein catabolic process, regulation of cell migration, positive regulation of cell migration,regulation of microtubule polymerization or depolymerization, negative regulation of microtubule polymerization or depolymerization, regulation of microtubule depolymerization, positive regulation of protein complex assembly,response to nutrient levels, cellular response to extracellular stimulus, cellular response to nutrient levels, regulation of cellular protein metabolic process, negative regulation of cellular protein metabolic process, lysosome localization,protein modification by small protein conjugation, regulation of microtubule-based process, regulation of organelle organization, regulation of steroid hormone receptor signaling pathway, cellular response to stress, cellular response to oxidative stress, cellular protein localization, cellular response to reactive oxygen species, peptidyl-lysine deacetylation, regulation of locomotion, positive regulation of locomotion, response to hydrogen peroxide, response to starvation, regulation of apoptosis, positive regulation of apoptosis, regulation of programmed cell death, positive regulation of programmed cell death, negative regulation of catalytic activity, negative regulation of protein complex disassembly, regulation of protein complex disassembly, regulation of protein complex assembly, regulation of cellular component biogenesis, negative regulation of molecular function, cellular protein catabolic process, cellular macromolecule catabolic process, establishment of protein localization, regulation of transcription, negative regulation of proteolysis, intracellular transport, negative regulation of cellular component organization, positive regulation of cellular component organization, negative regulation of protein metabolic process, regulation of RNA metabolic process, regulation of cell motion, positive regulation of cell motion, chromosome organization, regulation of oxidoreductase activity, negative regulation of oxidoreductase activity, regulation of cytoskeleton organization,negative regulation of cytoskeleton organization, proteolysis involved in cellular protein catabolic process, organelle localization, response to misfolded protein, response to protein stimulus, regulation of microtubule-based movement,regulation of androgen receptor signaling pathway, cellular response to hydrogen peroxide, regulation of microtubule cytoskeleton organization, protein modification by small protein conjugation or removal, cellular macromolecule localization, regulation of oxygen and reactive oxygen species metabolic process,
show all
Cellular Localization:
Cytoplasm . Cytoplasm, cytoskeleton . Nucleus . Perikaryon . Cell projection, dendrite . Cell projection, axon . It is mainly cytoplasmic, where it is associated with microtubules. .
show all
Signaling Pathway
Reference Citation({{totalcount}})
Catalog: KA3290C
Size
Price
Status
Qty.
96well
$330.00
In stock
0
Add to cart


Collected


Collect
Recently Viewed Products
Clear allPRODUCTS
CUSTOMIZED
ABOUT US
Toggle night Mode
{{pinfoXq.title || ''}}
Catalog: {{pinfoXq.catalog || ''}}
Filter:
All
{{item.name}}
{{pinfo.title}}
-{{pinfo.catalog}}
Main Information
Target
{{pinfo.target}}
Reactivity
{{pinfo.react}}
Applications
{{pinfo.applicat}}
Conjugate/Modification
{{pinfo.coupling}}/{{pinfo.modific}}
MW (kDa)
{{pinfo.mwcalc}}
Host Species
{{pinfo.hostspec}}
Isotype
{{pinfo.isotype}}
Product {{index}}/{{pcount}}
Prev
Next
{{pvTitle}}
Scroll wheel zooms the picture
{{pvDescr}}








.jpg)