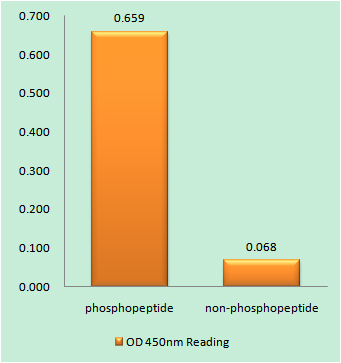
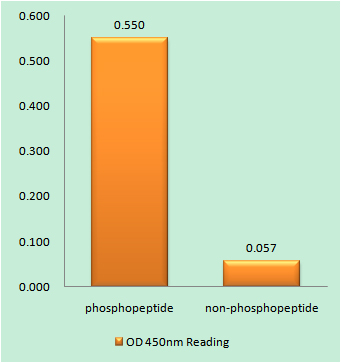
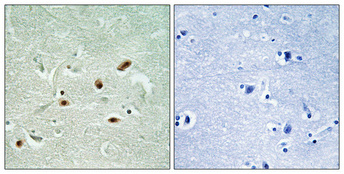
Catalog: KA3244C
Size
Price
Status
Qty.
96well
$330.00
In stock
0
Add to cart


Collected


Collect
Main Information
Reactivity
Human, Mouse
Applications
ELISA
Conjugate/Modification
Unmodified
Detailed Information
Storage
2-8°C/6 months,Ship by ice bag
Modification
Unmodified
Detection Method
Colorimetric
Related Products
Antigen&Target Information
Gene Name:
PRKDC
show all
Other Name:
DNA-dependent protein kinase catalytic subunit ;
DNA-PK catalytic subunit ;
DNA-PKcs ;
DNPK1 ;
p460 ;
DNA-PK catalytic subunit ;
DNA-PKcs ;
DNPK1 ;
p460 ;
show all
Background:
catalytic activity:ATP + a protein = ADP + a phosphoprotein.,enzyme regulation:Inhibited by wortmannin. Activity of the enzyme seems to be attenuated by autophosphorylation.,function:Serine/threonine-protein kinase that acts as a molecular sensor for DNA damage. Involved in DNA nonhomologous end joining (NHEJ) required for double-strand break (DSB) repair and V(D)J recombination. Must be bound to DNA to express its catalytic properties. Promotes processing of hairpin DNA structures in V(D)J recombination by activation of the hairpin endonuclease artemis (DCLRE1C). The assembly of the DNA-PK complex at DNA ends is also required for the NHEJ ligation step. Required to protect and align broken ends of DNA. May also act as a scaffold protein to aid the localization of DNA repair proteins to the site of damage. Found at the ends of chromosomes, suggesting a further role in the maintenance of telomeric stability and the prevention of chromosomal end fusion. Also involved in modulation of transcription. Recognizes the substrate consensus sequence [ST]-Q. Phosphorylates 'Ser-139' of histone variant H2AX/H2AFX, thereby regulating DNA damage response mechanism. Phosphorylates DCLRE1C, c-Abl/ABL1, histone H1, HSPCA, c-jun/JUN, p53/TP53, PARP1, POU2F1, DHX9, SRF, XRCC1, XRCC1, XRCC4, XRCC5, XRCC6, WRN, c-myc/MYC and RFA2. Can phosphorylate C1D not only in the presence of linear DNA but also in the presence of supercoiled DNA. Ability to phosphorylate TP53/p53 in the presence of supercoiled DNA is dependent on C1D.,PTM:Phosphorylated upon DNA damage, probably by ATM or ATR. Autophosphorylated on Thr-2609, Thr-2638 and Thr-2647. Thr-2609 is a DNA damage-inducible phosphorylation site (inducible with ionizing radiation, IR). Autophosphorylation induces a conformational change that leads to remodeling of the DNA-PK complex, requisite for efficient end processing and DNA repair.,similarity:Belongs to the PI3/PI4-kinase family.,similarity:Contains 1 FAT domain.,similarity:Contains 1 FATC domain.,similarity:Contains 1 PI3K/PI4K domain.,similarity:Contains 2 HEAT repeats.,similarity:Contains 3 TPR repeats.,subunit:DNA-PK is a heterotrimer of PRKDC and the Ku p70-p86 (XRCC6-XRCC5) dimer. Formation of this complex may be promoted by interaction with ILF3. Associates with the DNA-bound Ku heterodimer, but it can also bind to and be activated by free DNA. Interacts with DNA-PKcs-interacting protein (KIP) with the region upstream the kinase domain. PRKDC alone also interacts with and phosphorylates DCLRE1C, thereby activating the latent endonuclease activity of this protein. Interacts with C1D.,
show all
Function:
telomere maintenance, non-recombinational repair, somitogenesis, cell activation, somatic diversification of immune receptors, hemopoietic progenitor cell differentiation, immune effector process, lymphoid progenitor cell differentiation,B cell lineage commitment, pro-B cell differentiation, T cell lineage commitment, immunoglobulin production, production of molecular mediator of immune response, immune system development, leukocyte differentiation, somatic diversification of immune receptors via germline recombination within a single locus, somatic diversification of T cell receptor genes, somatic recombination of T cell receptor gene segments, regionalization, reproductive developmental process, DNA metabolic process, DNA repair, double-strand break repair, double-strand break repair via nonhomologous end joining, DNA recombination, regulation of transcription, DNA-dependent, regulation of transcription from RNA polymerase II promoter, protein amino acid phosphorylation, phosphorus metabolic process,phosphate metabolic process, cell motion, immune response, response to DNA damage stimulus, gamete generation,pattern specification process, heart development, cell death, germ cell migration, response to radiation, response to abiotic stimulus, response to endogenous stimulus, response to hormone stimulus, embryonic development ending in birth or egg hatching, positive regulation of biosynthetic process, anterior/posterior pattern formation, response to organic substance, response to ionizing radiation, response to gamma radiation, regulation of specific transcription from RNA polymerase II promoter, positive regulation of specific transcription from RNA polymerase II promoter,positive regulation of macromolecule biosynthetic process, positive regulation of macromolecule metabolic process,posttranscriptional regulation of gene expression, developmental programmed cell death, positive regulation of gene expression, regulation of cell death, positive regulation of cell death, programmed cell death, death, phosphorylation,somatic cell DNA recombination, somatic diversification of immunoglobulins, somatic recombination of immunoglobulin gene segments, cell migration, peptidyl-serine phosphorylation, peptidyl-serine modification, sexual reproduction,hemopoiesis, lymphocyte differentiation, B cell differentiation, T cell differentiation, positive regulation of cellular biosynthetic process, regulation of protein stability, protein destabilization, telomere organization, multicellular organism reproduction, regulation of gene-specific transcription, response to insulin stimulus, cellular response to insulin stimulus, cellular response to hormone stimulus, T cell differentiation in the thymus, V(D)J recombination,immunoglobulin V(D)J recombination, T cell receptor V(D)J recombination, cellular response to stress, germ cell programmed cell death, segmentation, T cell activation, B cell activation, homeostatic process, regulation of apoptosis,chordate embryonic development, positive regulation of apoptosis, regulation of programmed cell death, positive regulation of programmed cell death, positive regulation of gene-specific transcription, response to peptide hormone stimulus, cell fate commitment, leukocyte activation, regulation of transcription, positive regulation of transcription, DNA-dependent, positive regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process, positive regulation of transcription, positive regulation of transcription from RNA polymerase II promoter, lymphocyte activation, hemopoietic or lymphoid organ development, reproductive process in a multicellular organism, reproductive cellular process, cell motility, positive regulation of nitrogen compound metabolic process, regulation of RNA metabolic process, positive regulation of RNA metabolic process, chromosome organization, localization of cell, anatomical structure homeostasis,
show all
Cellular Localization:
Nucleus . Nucleus, nucleolus .
show all
Signaling Pathway
Reference Citation({{totalcount}})
Catalog: KA3244C
Size
Price
Status
Qty.
96well
$330.00
In stock
0
Add to cart


Collected


Collect
Recently Viewed Products
Clear allPRODUCTS
CUSTOMIZED
ABOUT US
Toggle night Mode
{{pinfoXq.title || ''}}
Catalog: {{pinfoXq.catalog || ''}}
Filter:
All
{{item.name}}
{{pinfo.title}}
-{{pinfo.catalog}}
Main Information
Target
{{pinfo.target}}
Reactivity
{{pinfo.react}}
Applications
{{pinfo.applicat}}
Conjugate/Modification
{{pinfo.coupling}}/{{pinfo.modific}}
MW (kDa)
{{pinfo.mwcalc}}
Host Species
{{pinfo.hostspec}}
Isotype
{{pinfo.isotype}}
Product {{index}}/{{pcount}}
Prev
Next
{{pvTitle}}
Scroll wheel zooms the picture
{{pvDescr}}