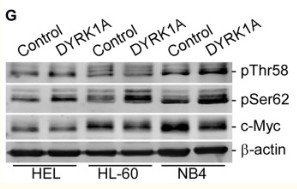
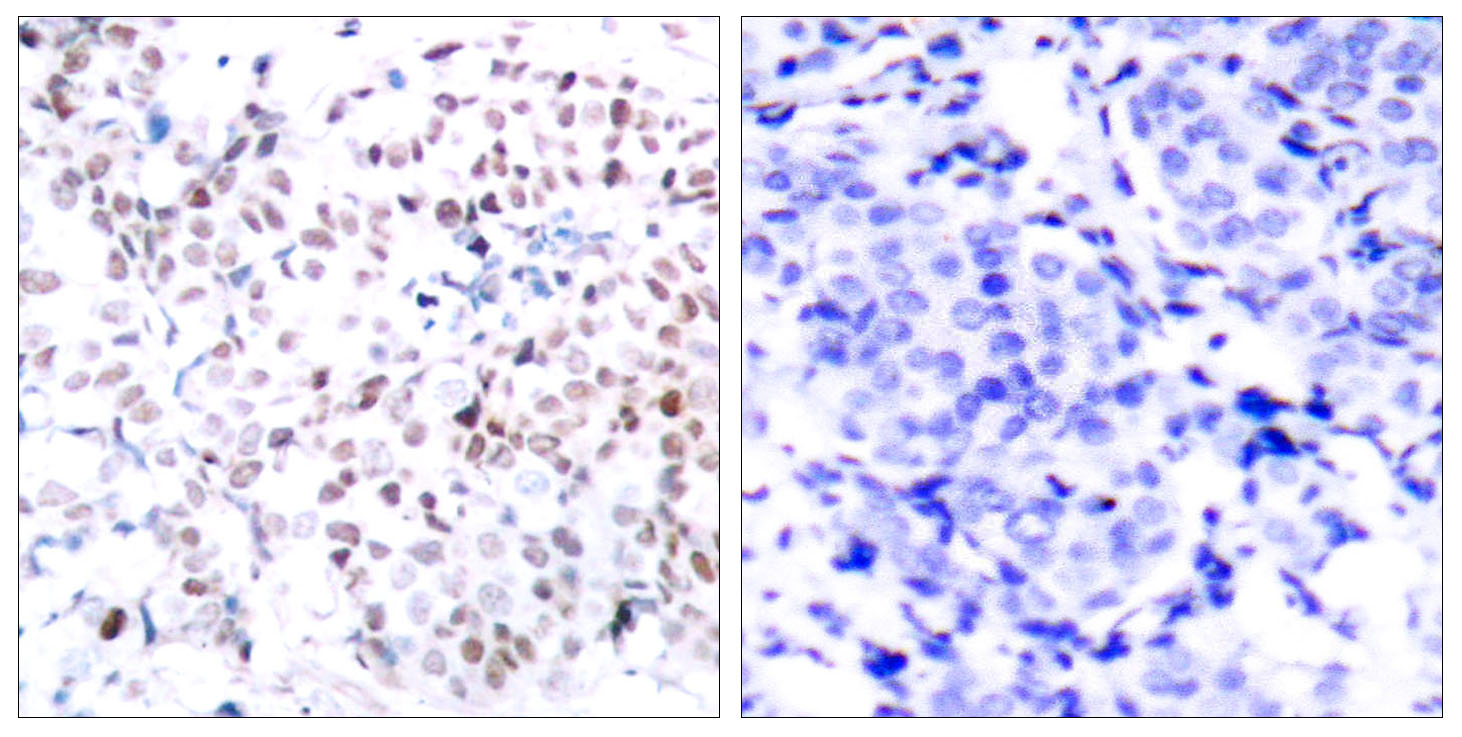
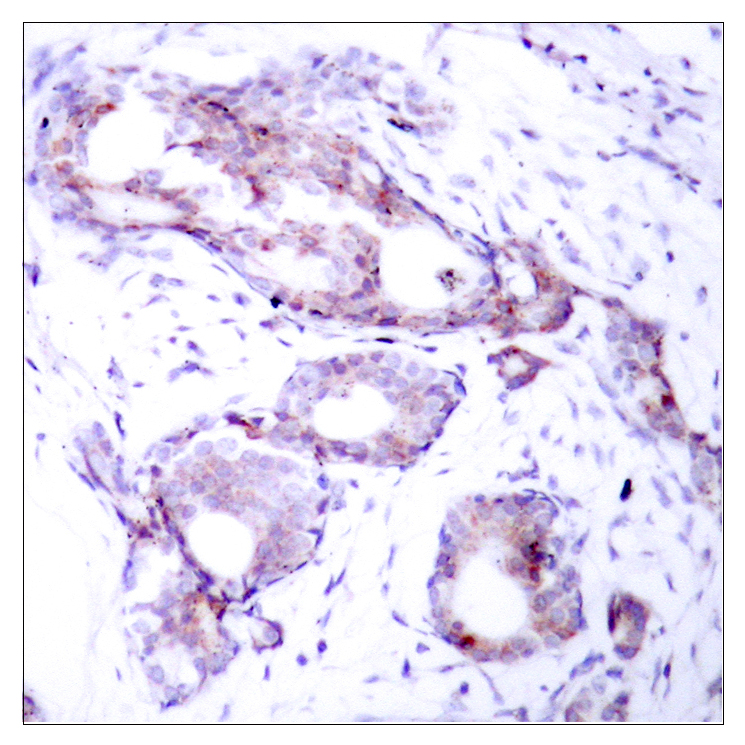
Catalog: KA3149C
Size
Price
Status
Qty.
96well
$330.00
In stock
0
Add to cart


Collected


Collect
Main Information
Reactivity
Human, Mouse, Rat
Applications
ELISA
Conjugate/Modification
Unmodified
Detailed Information
Storage
2-8°C/6 months,Ship by ice bag
Modification
Unmodified
Detection Method
Colorimetric
Related Products
Antigen&Target Information
Gene Name:
MYC BHLHE39
show all
Other Name:
MYC ;
BHLHE39 ;
Myc proto-oncogene protein ;
Class E basic helix-loop-helix protein 39 ;
bHLHe39 ;
Proto-oncogene c-Myc ;
Transcription factor p64
BHLHE39 ;
Myc proto-oncogene protein ;
Class E basic helix-loop-helix protein 39 ;
bHLHe39 ;
Proto-oncogene c-Myc ;
Transcription factor p64
show all
Database Link:
Background:
disease:A chromosomal aberration involving MYC may be a cause of a form of B-cell chronic lymphocytic leukemia. Translocation t(8;12)(q24;q22) with BTG1.,disease:Overexpression of MYC is implicated in the etiology of a variety of hematopoietic tumors.,function:Participates in the regulation of gene transcription. Binds DNA both in a non-specific manner and also specifically to recognizes the core sequence 5'-CAC[GA]TG-3'. Seems to activate the transcription of growth-related genes.,online information:Myc entry,PTM:Phosphorylated by PRKDC.,similarity:Contains 1 basic helix-loop-helix (bHLH) domain.,subunit:Efficient DNA binding requires dimerization with another bHLH protein. Binds DNA as a heterodimer with MAX. Interacts with TAF1C and SPAG9. Interacts with PARP10. Interacts with KDM5A and KDM5B.,
show all
Function:
DNA catabolic process, endonucleolytic, skeletal system development, B cell apoptosis, release of cytochrome c from mitochondria, regulation of B cell apoptosis, positive regulation of B cell apoptosis, monosaccharide metabolic process,glucose metabolic process, DNA metabolic process, DNA catabolic process, DNA fragmentation involved in apoptosis,transcription, transcription, DNA-dependent, transcription initiation, regulation of transcription, DNA-dependent,regulation of transcription from RNA polymerase II promoter, transcription from RNA polymerase II promoter, protein complex assembly, cellular ion homeostasis, cellular iron ion homeostasis, apoptosis, anti-apoptosis, induction of apoptosis, activation of caspase activity, cell structure disassembly during apoptosis, nucleus organization,mitochondrion organization, cell cycle, cell cycle arrest, regulation of mitotic cell cycle, sensory organ development,sensory perception, sensory perception of sound, cell death, cell proliferation, positive regulation of cell proliferation,induction of apoptosis by intracellular signals, activation of pro-apoptotic gene products, negative regulation of survival gene product expression, apoptotic mitochondrial changes, macromolecule catabolic process, response to radiation, detection of external stimulus, detection of abiotic stimulus, response to mechanical stimulus, response to abiotic stimulus, positive regulation of biosynthetic process, response to organic substance, positive regulation of macromolecule biosynthetic process, positive regulation of macromolecule metabolic process, negative regulation of macromolecule metabolic process, positive regulation of gene expression, negative regulation of gene expression,regulation of cell death, positive regulation of cell death, positive regulation of peptidase activity, programmed cell death, induction of programmed cell death, death, protein processing, hexose metabolic process, cellular homeostasis,cell cycle process, cellular component disassembly, cellular cation homeostasis, cellular di-, tri-valent inorganic cation homeostasis, apoptotic nuclear changes, positive regulation of cellular biosynthetic process, regulation of telomere maintenance, RNA biosynthetic process, regulation of homeostatic process, regulation of organelle organization,regulation of chromosome organization, regulation of cell proliferation, ear morphogenesis, middle ear morphogenesis,homeostatic process, regulation of apoptosis, positive regulation of apoptosis, negative regulation of apoptosis,regulation of programmed cell death, positive regulation of programmed cell death, negative regulation of programmed cell death, positive regulation of catalytic activity, response to alkaloid, positive regulation of caspase activity, regulation of caspase activity, pigmentation, ear development, macromolecular complex subunit organization,positive regulation of molecular function, cellular macromolecule catabolic process, regulation of transcription,regulation of survival gene product expression, positive regulation of transcription, DNA-dependent, positive regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process, positive regulation of transcription, positive regulation of transcription from RNA polymerase II promoter, embryonic organ morphogenesis,embryonic organ development, embryonic morphogenesis, skeletal system morphogenesis, chemical homeostasis, ion homeostasis, neurological system process, cognition, detection of stimulus involved in sensory perception, detection of mechanical stimulus involved in sensory perception of sound, sensory perception of mechanical stimulus, detection of mechanical stimulus involved in sensory perception, detection of mechanical stimulus, regulation of DNA metabolic process, positive regulation of nitrogen compound metabolic process, regulation of RNA metabolic process, positive regulation of RNA metabolic process, regulation of hydrolase activity, positive regulation of hydrolase activity, protein maturation, detection of stimulus, regulation of cell cycle, regulation of peptidase activity, regulation of endopeptidase activity, di-, tri-valent inorganic cation homeostasis, iron ion homeostasis, cation homeostasis, cellular chemical homeostasis, negative regulation of cell death, macromolecular complex assembly, lymphocyte apoptosis, regulation of lymphocyte apoptosis, positive regulation of lymphocyte apoptosis, protein complex biogenesis,
show all
Cellular Localization:
Nucleus, nucleoplasm . Nucleus, nucleolus .
show all
Signaling Pathway
Cellular Processes >> Cell growth and death >> Cell cycle
Cellular Processes >> Cell growth and death >> Cellular senescence
Cellular Processes >> Cellular community - eukaryotes >> Signaling pathways regulating pluripotency of stem cells
Organismal Systems >> Endocrine system >> Thyroid hormone signaling pathway
Human Diseases >> Cancer: overview >> Pathways in cancer
Human Diseases >> Cancer: overview >> Transcriptional misregulation in cancer
Human Diseases >> Cancer: overview >> MicroRNAs in cancer
Human Diseases >> Cancer: overview >> Central carbon metabolism in cancer
Human Diseases >> Cancer: specific types >> Colorectal cancer
Human Diseases >> Cancer: specific types >> Hepatocellular carcinoma
Human Diseases >> Cancer: specific types >> Gastric cancer
Human Diseases >> Cancer: specific types >> Thyroid cancer
Human Diseases >> Cancer: specific types >> Acute myeloid leukemia
Human Diseases >> Cancer: specific types >> Chronic myeloid leukemia
Human Diseases >> Cancer: specific types >> Bladder cancer
Human Diseases >> Cancer: specific types >> Endometrial cancer
Human Diseases >> Cancer: specific types >> Breast cancer
Human Diseases >> Cancer: specific types >> Small cell lung cancer
Environmental Information Processing >> Signal transduction >> MAPK signaling pathway
Environmental Information Processing >> Signal transduction >> ErbB signaling pathway
Environmental Information Processing >> Signal transduction >> Wnt signaling pathway
Environmental Information Processing >> Signal transduction >> TGF-beta signaling pathway
Environmental Information Processing >> Signal transduction >> Hippo signaling pathway
Environmental Information Processing >> Signal transduction >> JAK-STAT signaling pathway
Environmental Information Processing >> Signal transduction >> PI3K-Akt signaling pathway
Reference Citation({{totalcount}})
Catalog: KA3149C
Size
Price
Status
Qty.
96well
$330.00
In stock
0
Add to cart


Collected


Collect
Recently Viewed Products
Clear allPRODUCTS
CUSTOMIZED
ABOUT US
Toggle night Mode
{{pinfoXq.title || ''}}
Catalog: {{pinfoXq.catalog || ''}}
Filter:
All
{{item.name}}
{{pinfo.title}}
-{{pinfo.catalog}}
Main Information
Target
{{pinfo.target}}
Reactivity
{{pinfo.react}}
Applications
{{pinfo.applicat}}
Conjugate/Modification
{{pinfo.coupling}}/{{pinfo.modific}}
MW (kDa)
{{pinfo.mwcalc}}
Host Species
{{pinfo.hostspec}}
Isotype
{{pinfo.isotype}}
Product {{index}}/{{pcount}}
Prev
Next
{{pvTitle}}
Scroll wheel zooms the picture
{{pvDescr}}