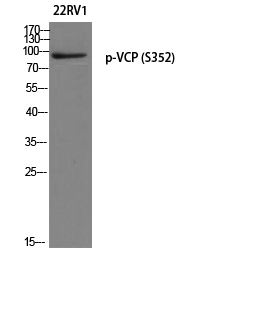
Catalog: KA1765C
Size
Price
Status
Qty.
96well
$470.00
In stock
0
Add to cart


Collected


Collect
Main Information
Reactivity
Human, Mouse, Rat
Applications
ELISA
Conjugate/Modification
Phospho
Detailed Information
Storage
2-8°C/6 months,Ship by ice bag
Modification
Phospho
Detection Method
Colorimetric
Related Products
Antigen&Target Information
Gene Name:
VCP
show all
Other Name:
Transitional endoplasmic reticulum ATPase ;
TER ATPase ;
15S Mg ;
2+ ;
-ATPase p97 subunit ;
Valosin-containing protein ;
VCP ;
TER ATPase ;
15S Mg ;
2+ ;
-ATPase p97 subunit ;
Valosin-containing protein ;
VCP ;
show all
Background:
disease:Defects in VCP are the cause of inclusion body myopathy with early-onset Paget disease and frontotemporal dementia (IBMPFD) [MIM:167320]; also known as muscular dystrophy, limb-girdle, with Paget disease of bone or pagetoid amyotrophic lateral sclerosis or pagetoid neuroskeletal syndrome or lower motor neuron degeneration with Paget-like bone disease. IBMPFD features adult-onset proximal and distal muscle weakness (clinically resembling limb girdle muscular dystrophy), early-onset Paget disease of bone in most cases and premature frontotemporal dementia.,function:Necessary for the fragmentation of Golgi stacks during mitosis and for their reassembly after mitosis. Involved in the formation of the transitional endoplasmic reticulum (tER). The transfer of membranes from the endoplasmic reticulum to the Golgi apparatus occurs via 50-70 nm transition vesicles which derive from part-rough, part-smooth transitional elements of the endoplasmic reticulum (tER). Vesicle budding from the tER is an ATP-dependent process. The ternary complex containing UFD1L, VCP and NPLOC4 binds ubiquitinated proteins and is necessary for the export of misfolded proteins from the ER to the cytoplasm, where they are degraded by the proteasome. The NPLOC4-UFD1L-VCP complex regulates spindle disassembly at the end of mitosis and is necessary for the formation of a closed nuclear envelope (By similarity). Regulates E3 ubiquitin-protein ligase activity of RNF19A.,PTM:Phosphorylated by tyrosine kinases in response to T-cell antigen receptor activation (By similarity). Phosphorylated upon DNA damage, probably by ATM or ATR.,similarity:Belongs to the AAA ATPase family.,subcellular location:Present in the neuronal hyaline inclusion bodies specifically found in motor neurons from amyotrophic lateral sclerosis patients. Present in the Lewy bodies specifically found in neurons from Parkinson disease patients.,subunit:Homohexamer. Forms a ring-shaped particle of 12.5 nm diameter, that displays 6-fold radial symmetry. Part of a ternary complex containing STX5A, NSFL1C and VCP. NSFL1C forms a homotrimer that binds to one end of a VCP homohexamer. The complex binds to membranes enriched in phosphatidylethanolamine-containing lipids and promotes Golgi membrane fusion. Binds to a heterodimer of NPLOC4 and UFD1L, binding to this heterodimer inhibits Golgi-membrane fusion. Interaction with VCIP135 leads to dissociation of the complex via ATP hydrolysis by VCP. Part of a ternary complex containing NPLOC4, UFD1L and VCP. Interacts with NSFL1C-like protein p37; the complex has membrane fusion activity and is required for Golgi and endoplasmic reticulum biogenesis (By similarity). Interacts with SELS/VIMP and SYVN1, as well as with DERL1, DERL2 and DERL3; which probably transfer misfolded proteins from the ER to VCP. Interacts with SVIP. Component of a complex required to couple retrotranslocation, ubiquitination and deglycosylation composed of NGLY1, SAKS1, AMFR, VCP and RAD23B. Directly interacts with UBXD2 and RNF19A. Interacts with CASR. Interacts with UBXN6 and UBE4B.,
show all
Function:
DNA metabolic process, DNA repair, double-strand break repair, protein complex assembly, proteolysis, ubiquitin-dependent protein catabolic process, intracellular protein transport, ER to Golgi vesicle-mediated transport, apoptosis,activation of caspase activity, response to DNA damage stimulus, ER-nuclear signaling pathway, response to unfolded protein, protein localization, cell death, macromolecule catabolic process, response to organic substance, proteasomal protein catabolic process, regulation of cell death, positive regulation of peptidase activity, programmed cell death,protein transport, vesicle-mediated transport, death, protein ubiquitination, modification-dependent protein catabolic process, protein catabolic process, ER-associated protein catabolic process, endoplasmic reticulum unfolded protein response, retrograde protein transport, ER to cytosol, protein modification by small protein conjugation, cellular response to stress, cellular protein localization, cellular response to unfolded protein, response to endoplasmic reticulum stress, regulation of apoptosis, regulation of programmed cell death, positive regulation of catalytic activity,proteasomal ubiquitin-dependent protein catabolic process, positive regulation of caspase activity, regulation of caspase activity, modification-dependent macromolecule catabolic process, macromolecular complex subunit organization, positive regulation of molecular function, cellular protein catabolic process, cellular macromolecule catabolic process, establishment of protein localization, intracellular transport, Golgi vesicle transport, protein oligomerization, protein homooligomerization, regulation of hydrolase activity, positive regulation of hydrolase activity,proteolysis involved in cellular protein catabolic process, response to protein stimulus, regulation of peptidase activity,regulation of endopeptidase activity, macromolecular complex assembly, protein complex biogenesis, protein modification by small protein conjugation or removal, cellular macromolecule localization,
show all
Cellular Localization:
Cytoplasm, cytosol . Endoplasmic reticulum . Nucleus . Cytoplasm, Stress granule . Present in the neuronal hyaline inclusion bodies specifically found in motor neurons from amyotrophic lateral sclerosis patients (PubMed:15456787). Present in the Lewy bodies specifically found in neurons from Parkinson disease patients (PubMed:15456787). Recruited to the cytoplasmic surface of the endoplasmic reticulum via interaction with AMFR/gp78 (PubMed:16168377). Following DNA double-strand breaks, recruited to the sites of damage (PubMed:22120668). Recruited to stalled replication forks via interaction with SPRTN (PubMed:23042605). Recruited to damaged lysosomes decorated with K48-linked ubiquitin chains (PubMed:27753622). Colocalizes with TIA1, ZFAND1 and G3BP1 in cytoplasmic stress granules (SGs) in response to arsenite-induced stress treatment (PubMed:29804830). .
show all
Signaling Pathway
Cellular Processes >> Transport and catabolism >> Mitophagy - animal
Human Diseases >> Neurodegenerative disease >> Amyotrophic lateral sclerosis
Human Diseases >> Neurodegenerative disease >> Pathways of neurodegeneration - multiple diseases
Genetic Information Processing >> Folding, sorting and degradation >> Protein processing in endoplasmic reticulum
Reference Citation({{totalcount}})
Catalog: KA1765C
Size
Price
Status
Qty.
96well
$470.00
In stock
0
Add to cart


Collected


Collect
Recently Viewed Products
Clear allPRODUCTS
CUSTOMIZED
ABOUT US
Toggle night Mode
{{pinfoXq.title || ''}}
Catalog: {{pinfoXq.catalog || ''}}
Filter:
All
{{item.name}}
{{pinfo.title}}
-{{pinfo.catalog}}
Main Information
Target
{{pinfo.target}}
Reactivity
{{pinfo.react}}
Applications
{{pinfo.applicat}}
Conjugate/Modification
{{pinfo.coupling}}/{{pinfo.modific}}
MW (kDa)
{{pinfo.mwcalc}}
Host Species
{{pinfo.hostspec}}
Isotype
{{pinfo.isotype}}
Product {{index}}/{{pcount}}
Prev
Next
{{pvTitle}}
Scroll wheel zooms the picture
{{pvDescr}}