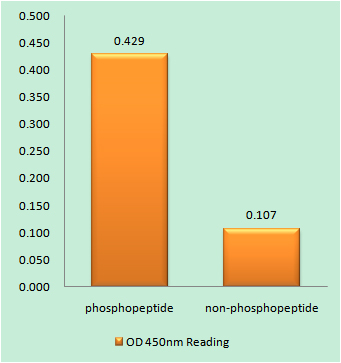
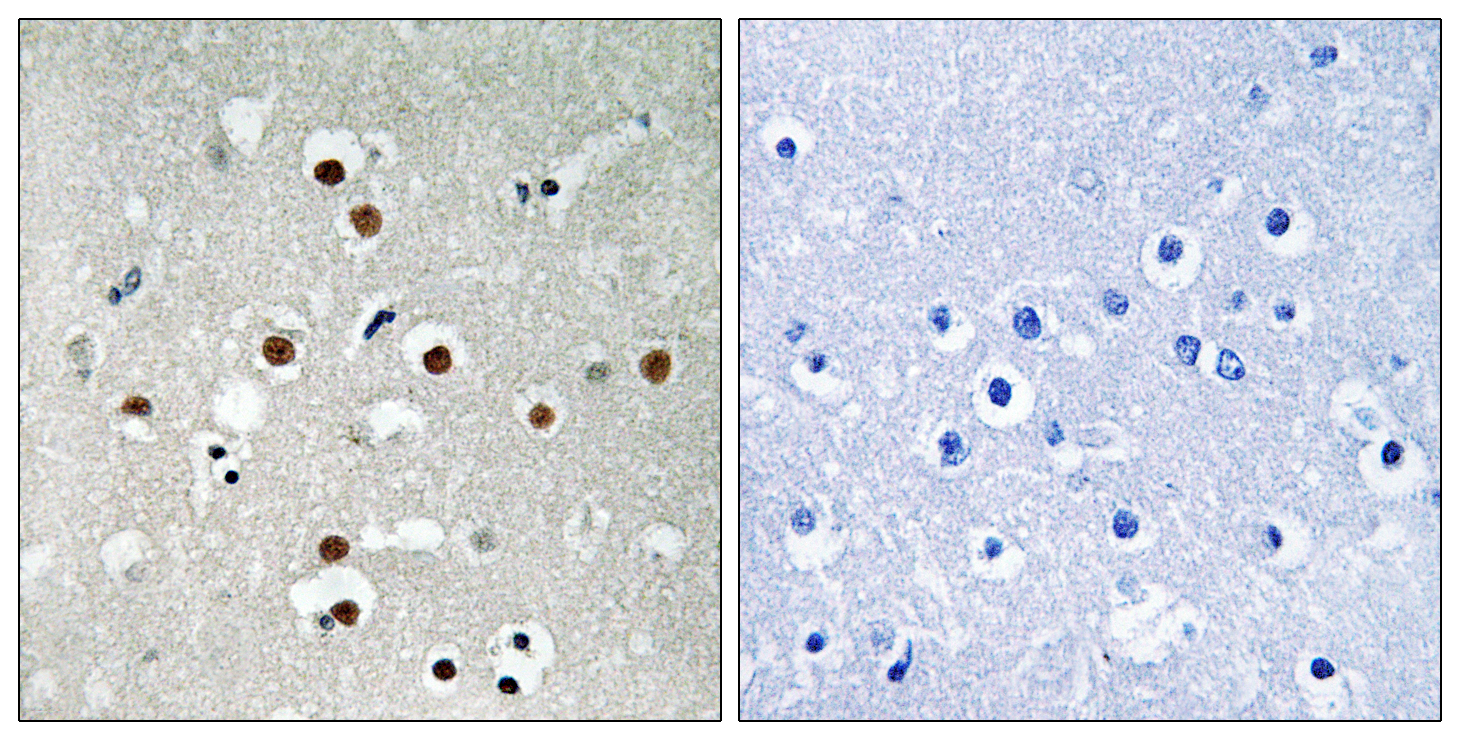
Catalog: KA1753C
Size
Price
Status
Qty.
96well
$470.00
In stock
0
Add to cart


Collected


Collect
Main Information
Reactivity
Human, Mouse
Applications
ELISA
Conjugate/Modification
Phospho
Detailed Information
Storage
2-8°C/6 months,Ship by ice bag
Modification
Phospho
Detection Method
Colorimetric
Related Products
Antigen&Target Information
Gene Name:
MEF2C
show all
Other Name:
Myocyte-specific enhancer factor 2C
show all
Background:
alternative products:Additional isoforms seem to exist,developmental stage:Expression is highest during the early stages of postnatal development, at later stages levels greatly decrease.,domain:The beta domain, missing in a number of isoforms, is required for enhancement of transcriptional activity.,function:Transcription activator which binds specifically to the MEF2 element present in the regulatory regions of many muscle-specific genes. Controls cardiac morphogenesis and myogenesis, and is also involved in vascular development. May also be involved in neurogenesis and in the development of cortical architecture (By similarity). Isoform 3 and isoform 4, which lack the repressor domain, are more active than isoform 1 and isoform 2.,PTM:Acetylated by p300 on several sites in diffentiating myocytes. Acetylation on Lys-4 increases DNA binding and transactivation.,PTM:Phosphorylation on Ser-59 enhances DNA binding activity (By similarity). Phosphorylation on Ser-396 is required for Lys-391 sumoylation and inhibits transcriptional activity.,PTM:Proteolytically cleaved in cerebellar granule neurons, probably by caspase 7, following neurotoxicity. Preferentially cleaves the CDK5-mediated hyperphosphorylated form which leads to neuron apoptosis and transcriptional inactivation.,PTM:Sumoylated on Lys-391 by SUMO2 but not by SUMO1 represses transcriptional activity.,similarity:Belongs to the MEF2 family.,similarity:Contains 1 MADS-box domain.,similarity:Contains 1 Mef2-type DNA-binding domain.,subunit:Forms a complex with class II HDACs in undifferentiating cells. On myogenic differentiation, HDACs are released into the cytoplasm allowing MEF2s to interact with other proteins for activation. Interacts with EP300 in differentiating cells; the interaction acetylates MEF2C leading to increased DNA binding and activation. Interacts with HDAC7 and CARM1 (By similarity). Interacts with HDAC4, HDAC7 AND HDAC9; the interaction WITH HDACs represses transcriptional activity.,tissue specificity:Expressed in brain and skeletal muscle.,
show all
Function:
negative regulation of transcription from RNA polymerase II promoter, transcription, regulation of transcription, DNA-dependent, regulation of transcription from RNA polymerase II promoter, apoptosis, anti-apoptosis, muscle organ development, cell death, negative regulation of biosynthetic process, positive regulation of biosynthetic process,regulation of specific transcription from RNA polymerase II promoter, positive regulation of specific transcription from RNA polymerase II promoter, negative regulation of specific transcription from RNA polymerase II promoter, positive regulation of macromolecule biosynthetic process, negative regulation of macromolecule biosynthetic process, positive regulation of macromolecule metabolic process, negative regulation of macromolecule metabolic process, positive regulation of gene expression, negative regulation of gene expression, regulation of cell death, programmed cell death, death, negative regulation of transcription, negative regulation of cellular biosynthetic process, positive regulation of cellular biosynthetic process, negative regulation of gene-specific transcription, regulation of gene-specific transcription, regulation of apoptosis, negative regulation of apoptosis, regulation of programmed cell death,negative regulation of programmed cell death, positive regulation of gene-specific transcription, regulation of transcription, regulation of survival gene product expression, positive regulation of survival gene product expression,negative regulation of transcription, DNA-dependent, positive regulation of transcription, DNA-dependent, negative regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process, positive regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process, positive regulation of transcription, positive regulation of transcription from RNA polymerase II promoter, negative regulation of nitrogen compound metabolic process, positive regulation of nitrogen compound metabolic process, regulation of RNA metabolic process, negative regulation of RNA metabolic process, positive regulation of RNA metabolic process, negative regulation of cell death,
show all
Cellular Localization:
Nucleus . Cytoplasm, sarcoplasm .
show all
Tissue Expression:
Signaling Pathway
Organismal Systems >> Endocrine system >> Oxytocin signaling pathway
Human Diseases >> Cancer: overview >> Transcriptional misregulation in cancer
Environmental Information Processing >> Signal transduction >> MAPK signaling pathway
Environmental Information Processing >> Signal transduction >> Apelin signaling pathway
Environmental Information Processing >> Signal transduction >> cGMP-PKG signaling pathway
Reference Citation({{totalcount}})
Catalog: KA1753C
Size
Price
Status
Qty.
96well
$470.00
In stock
0
Add to cart


Collected


Collect
Recently Viewed Products
Clear allPRODUCTS
CUSTOMIZED
ABOUT US
Toggle night Mode
{{pinfoXq.title || ''}}
Catalog: {{pinfoXq.catalog || ''}}
Filter:
All
{{item.name}}
{{pinfo.title}}
-{{pinfo.catalog}}
Main Information
Target
{{pinfo.target}}
Reactivity
{{pinfo.react}}
Applications
{{pinfo.applicat}}
Conjugate/Modification
{{pinfo.coupling}}/{{pinfo.modific}}
MW (kDa)
{{pinfo.mwcalc}}
Host Species
{{pinfo.hostspec}}
Isotype
{{pinfo.isotype}}
Product {{index}}/{{pcount}}
Prev
Next
{{pvTitle}}
Scroll wheel zooms the picture
{{pvDescr}}