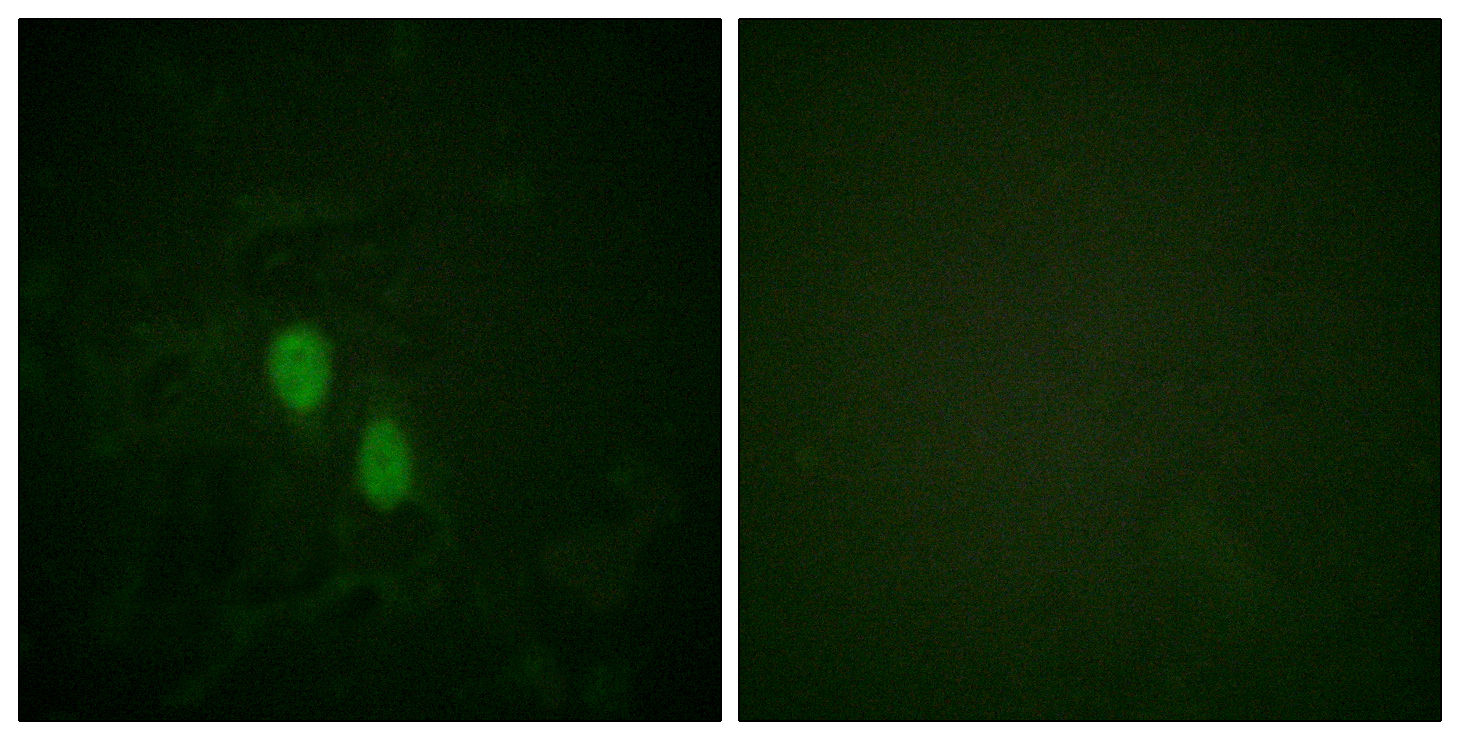
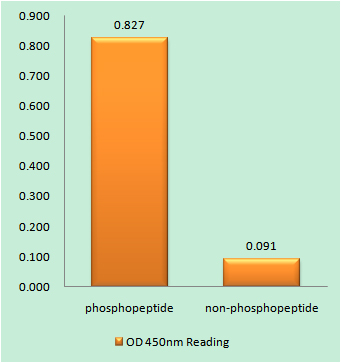
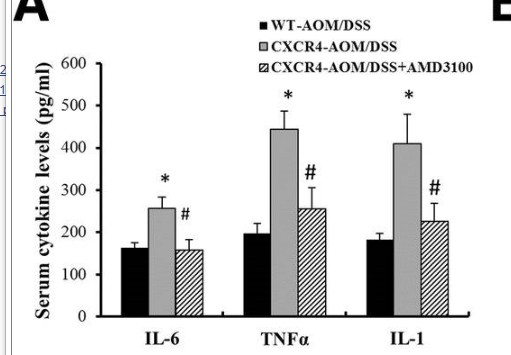
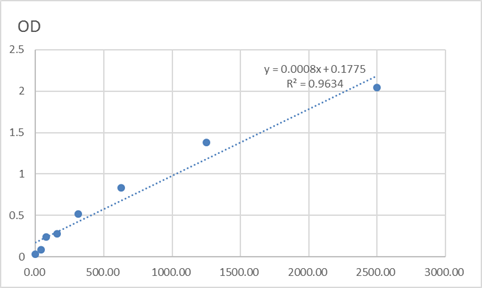
Catalog: KA1490C
Size
Price
Status
Qty.
96well
$470.00
In stock
0
Add to cart


Collected


Collect
Main Information
Reactivity
Human, Mouse, Rat
Applications
ELISA
Conjugate/Modification
Phospho
Detailed Information
Storage
2-8°C/6 months,Ship by ice bag
Modification
Phospho
Detection Method
Colorimetric
Related Products
Antigen&Target Information
Gene Name:
HNRNPD
show all
Other Name:
Heterogeneous nuclear ribonucleoprotein D0 ;
hnRNP D0 ;
AU-rich element RNA-binding protein 1 ;
hnRNP D0 ;
AU-rich element RNA-binding protein 1 ;
show all
Background:
function:Binds with high affinity to RNA molecules that contain AU-rich elements (AREs) found within the 3'-UTR of many proto-oncogenes and cytokine mRNAs. Also binds to double- and single-stranded DNA sequences in a specific manner and functions a transcription factor. Each of the RNA-binding domains specifically can bind solely to a single-stranded non-monotonous 5'-UUAG-3' sequence and also weaker to the single-stranded 5'-TTAGGG-3' telomeric DNA repeat. Binds RNA oligonucleotides with 5'-UUAGGG-3' repeats more tightly than the telomeric single-stranded DNA 5'-TTAGGG-3' repeats. Binding of RRM1 to DNA inhibits the formation of DNA quadruplex structure which may play a role in telomere elongation. May be involved in translationally coupled mRNA turnover. Implicated with other RNA-binding proteins in the cytoplasmic deadenylation/translational and decay interplay of the FOS mRNA mediated by the major coding-region determinant of instability (mCRD) domain.,PTM:Arg-345 is dimethylated, probably to asymmetric dimethylarginine.,sequence caution:Contaminating sequence. Sequence of unknown origin in the N-terminal part.,sequence caution:Several sequence conflicts.,similarity:Contains 2 RRM (RNA recognition motif) domains.,subcellular location:Component of ribonucleosomes.,subunit:Part of a complex associated with the FOS mCRD domain and consisting of PABPC1, PAIP1, CSDE1/UNR and SYNCRIP. Interacts with IGF2BP2.,
show all
Function:
RNA splicing, via transesterification reactions, RNA splicing, via transesterification reactions with bulged adenosine as nucleophile, nuclear mRNA splicing, via spliceosome, transcription, regulation of transcription, DNA-dependent, RNA processing, mRNA processing, RNA catabolic process, mRNA catabolic process, RNA splicing, macromolecule catabolic process, posttranscriptional regulation of gene expression, mRNA metabolic process, regulation of RNA stability,regulation of mRNA stability, RNA stabilization, cellular macromolecule catabolic process, regulation of transcription,mRNA stabilization, regulation of RNA metabolic process,
show all
Cellular Localization:
Nucleus. Cytoplasm. Localized in cytoplasmic mRNP granules containing untranslated mRNAs. Component of ribonucleosomes. Cytoplasmic localization oscillates diurnally.
show all
Reference Citation({{totalcount}})
Catalog: KA1490C
Size
Price
Status
Qty.
96well
$470.00
In stock
0
Add to cart


Collected


Collect
Recently Viewed Products
Clear allPRODUCTS
CUSTOMIZED
ABOUT US
Toggle night Mode
{{pinfoXq.title || ''}}
Catalog: {{pinfoXq.catalog || ''}}
Filter:
All
{{item.name}}
{{pinfo.title}}
-{{pinfo.catalog}}
Main Information
Target
{{pinfo.target}}
Reactivity
{{pinfo.react}}
Applications
{{pinfo.applicat}}
Conjugate/Modification
{{pinfo.coupling}}/{{pinfo.modific}}
MW (kDa)
{{pinfo.mwcalc}}
Host Species
{{pinfo.hostspec}}
Isotype
{{pinfo.isotype}}
Product {{index}}/{{pcount}}
Prev
Next
{{pvTitle}}
Scroll wheel zooms the picture
{{pvDescr}}