Catalog: KA1007C
Size
Price
Status
Qty.
96well
$470.00
In stock
0
Add to cart


Collected


Collect
Main Information
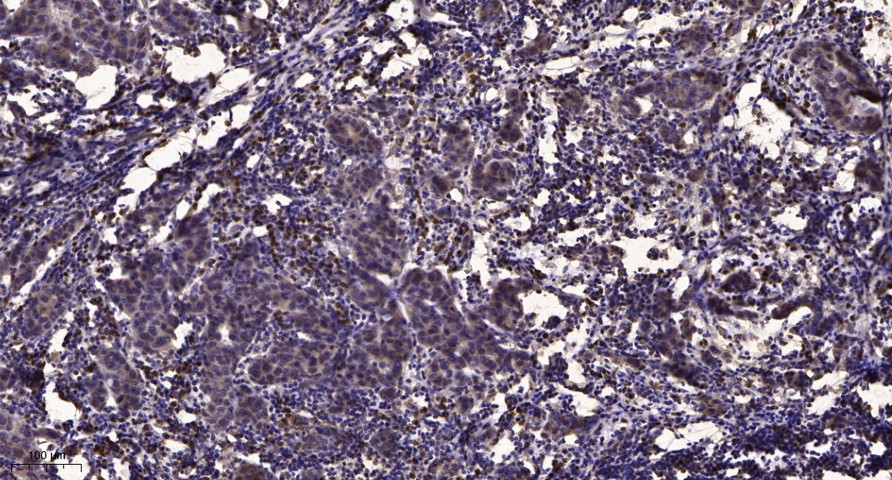
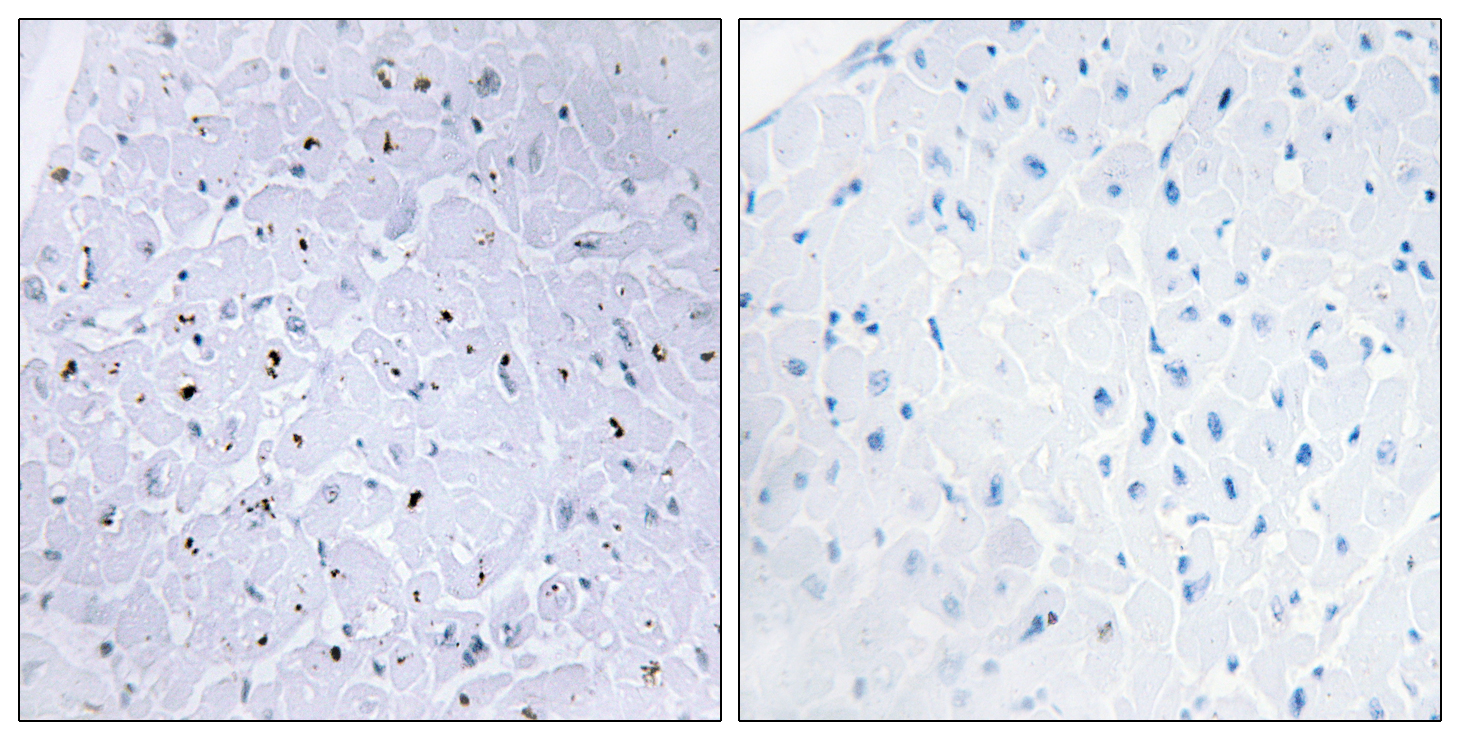
Target
53BP1 Phospho Ser6
Reactivity
Human, Mouse, Rat, Chicken(testedbyourcustomer)
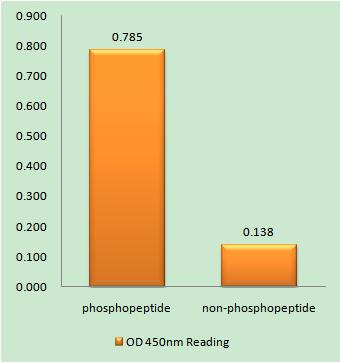
Applications
ELISA
Conjugate/Modification
Phospho
Detailed Information
Storage
2-8°C/6 months,Ship by ice bag
Modification
Phospho
Detection Method
Colorimetric
Related Products
Antigen&Target Information
Gene Name:
TP53BP1
show all
Protein Name:
Tumor suppressor p53-binding protein 1
show all
Other Name:
TP53BP1 ;
Tumor suppressor p53-binding protein 1 ;
53BP1 ;
p53-binding protein 1 ;
p53BP1
Tumor suppressor p53-binding protein 1 ;
53BP1 ;
p53-binding protein 1 ;
p53BP1
show all
Background:
This gene encodes a protein that functions in the DNA double-strand break repair pathway choice, promoting non-homologous end joining (NHEJ) pathways, and limiting homologous recombination. This protein plays multiple roles in the DNA damage response, including promoting checkpoint signaling following DNA damage, acting as a scaffold for recruitment of DNA damage response proteins to damaged chromatin, and promoting NHEJ pathways by limiting end resection following a double-strand break. These roles are also important during V(D)J recombination, class switch recombination and at unprotected telomeres. Alternative splicing results in multiple transcript variants encoding different isoforms. [provided by RefSeq, Aug 2017]
show all
Function:
Function:May have a role in checkpoint signaling during mitosis (By similarity). Enhances TP53-mediated transcriptional activation. Plays a role in the response to DNA damage.,PTM:Asymmetrically dimethylated on Arg residues by PRMT1. Methylation is required for DNA binding.,PTM:Phosphorylated at basal level in the absence of DNA damage. Hyper-phosphorylated in an ATM-dependent manner in response to DNA damage induced by ionizing radiation. Hyper-phosphorylated in an ATR-dependent manner in response to DNA damage induced by UV irradiation.,similarity:Contains 2 BRCT domains.,subcellular location:Associated with kinetochores. Both nuclear and cytoplasmic in some cells. Recruited to sites of DNA damage, such as double stand breaks. Methylation of histone H4 at 'Lys-20' is required for efficient localization to double strand breaks.,subunit:Interacts with IFI202A (By similarity). Binds to the central domain of TP53/p53. May form homo-oligomers. Interacts with DCLRE1C. Interacts with histone H2AFX and this requires phosphorylation of H2AFX on 'Ser-139'. Interacts with histone H4 that has been dimethylated at 'Lys-20'. Has low affinity for histone H4 containing monomethylated 'Lys-20'. Does not bind histone H4 containing unmethylated or trimethylated 'Lys-20'. Has low affinity for histone H3 that has been dimethylated on 'Lys-79'. Has very low affinity for histone H3 that has been monomethylated on 'Lys-79' (in vitro). Does not bind unmethylated histone H3.,
show all
Cellular Localization:
Nucleus
show all
Tissue Expression:
Research Areas:
>>NOD-like receptor signaling pathway
show all
Reference Citation({{totalcount}})
Catalog: KA1007C
Size
Price
Status
Qty.
96well
$470.00
In stock
0
Add to cart


Collected


Collect
Recently Viewed Products
Clear allPRODUCTS
CUSTOMIZED
ABOUT US
Toggle night Mode
{{pinfoXq.title || ''}}
Catalog: {{pinfoXq.catalog || ''}}
Filter:
All
{{item.name}}
{{pinfo.title}}
-{{pinfo.catalog}}
Main Information
Target
{{pinfo.target}}
Reactivity
{{pinfo.react}}
Applications
{{pinfo.applicat}}
Conjugate/Modification
{{pinfo.coupling}}/{{pinfo.modific}}
MW (kDa)
{{pinfo.mwcalc}}
Host Species
{{pinfo.hostspec}}
Isotype
{{pinfo.isotype}}
Product {{index}}/{{pcount}}
Prev
Next
{{pvTitle}}
Scroll wheel zooms the picture
{{pvDescr}}