PARP Monoclonal Antibody(M3)
- Catalog No.:YM3131
- Applications:WB
- Reactivity:Human;Chicken(testedbyourcustomer)
- Target:
- PARP
- Fields:
- >>Base excision repair;>>NF-kappa B signaling pathway;>>Apoptosis;>>Necroptosis;>>Diabetic cardiomyopathy
- Gene Name:
- PARP1
- Protein Name:
- Poly [ADP-ribose] polymerase 1
- Human Gene Id:
- 142
- Human Swiss Prot No:
- P09874
- Mouse Swiss Prot No:
- P11103
- Rat Gene Id:
- 25591
- Rat Swiss Prot No:
- P27008
- Immunogen:
- Synthetic Peptide of PARP
- Specificity:
- The antibody detects endogenous PARP protein.
- Formulation:
- PBS, pH 7.4, containing 0.5%BSA, 0.02% sodium azide as Preservative and 50% Glycerol.
- Source:
- Monoclonal, Mouse
- Dilution:
- WB 1:1000-3000
- Purification:
- The antibody was affinity-purified from mouse ascites by affinity-chromatography using specific immunogen.
- Storage Stability:
- -15°C to -25°C/1 year(Do not lower than -25°C)
- Other Name:
- Poly [ADP-ribose] polymerase 1 (PARP-1) (EC 2.4.2.30) (ADP-ribosyltransferase diphtheria toxin-like 1) (ARTD1) (NAD(+) ADP-ribosyltransferase 1) (ADPRT 1) (Poly[ADP-ribose] synthase 1)
- Observed Band(KD):
- 116kD
- Background:
- This gene encodes a chromatin-associated enzyme, poly(ADP-ribosyl)transferase, which modifies various nuclear proteins by poly(ADP-ribosyl)ation. The modification is dependent on DNA and is involved in the regulation of various important cellular processes such as differentiation, proliferation, and tumor transformation and also in the regulation of the molecular events involved in the recovery of cell from DNA damage. In addition, this enzyme may be the site of mutation in Fanconi anemia, and may participate in the pathophysiology of type I diabetes. [provided by RefSeq, Jul 2008],
- Function:
- catalytic activity:NAD(+) + (ADP-D-ribosyl)(n)-acceptor = nicotinamide + (ADP-D-ribosyl)(n+1)-acceptor.,function:Involved in the base excision repair (BER) pathway, by catalyzing the poly(ADP-ribosyl)ation of a limited number of acceptor proteins involved in chromatin architecture and in DNA metabolism. This modification follows DNA damages and appears as an obligatory step in a detection/signaling pathway leading to the reparation of DNA strand breaks.,miscellaneous:The ADP-D-ribosyl group of NAD(+) is transferred to an acceptor carboxyl group on a histone or the enzyme itself, and further ADP-ribosyl groups are transferred to the 2'-position of the terminal adenosine moiety, building up a polymer with an average chain length of 20-30 units.,PTM:Phosphorylated by PRKDC. Phosphorylated upon DNA damage, probably by ATM or ATR.,PTM:Poly-ADP-ribosylated by PARP2.,similarity:Contains 1 BRCT
- Subcellular Location:
- Nucleus . Nucleus, nucleolus . Chromosome . Localizes to sites of DNA damage. .
- Expression:
- Brain,Colon carcinoma,Fibroblast,Lung,Ovarian carcinoma,Skin,
Modulations of Keap1-Nrf2 signaling axis by TIIA ameliorated the oxidative stress-induced myocardial apoptosis. FREE RADICAL BIOLOGY AND MEDICINE Free Radical Bio Med. 2018 Feb;115:191 WB Mouse Neonatal rat ventricular myocytes (NRVMs)
The induction of apoptosis and autophagy in human hepatoma SMMC-7721 cells by combined treatment with vitamin C and polysaccharides extracted from Grifola frondosa. APOPTOSIS 2017 Sep 11 WB Human SMMC-7721 cell
Coptisine Induces Apoptosis in Human Hepatoma Cells Through Activating 67-kDa Laminin Receptor/cGMP Signaling. Frontiers in Pharmacology 2018 May 18 WB Human 1:500 SMMC7721 cell, HepG2 Cell
High CHMP4B expression is associated with accelerated cell proliferation and resistance to doxorubicin in hepatocellular carcinoma. TUMOR BIOLOGY Tumor Biol. 2015 Apr;36(4):2569-2581 WB Human HepG2 cell, Huh7 HCC cell
High expression of PINK1 promotes proliferation and chemoresistance of NSCLC. ONCOLOGY REPORTS Oncol Rep. 2017 Apr;37(4):2137-2146 WB Human NSCLC tissues A549 cell, H1299 cell
Effect of metformin on apoptosis, cell cycle arrest migration and invasion of A498 cells. Molecular Medicine Reports 2014 Mar 31 WB Human A498 cell
- June 19-2018
- WESTERN IMMUNOBLOTTING PROTOCOL
- June 19-2018
- IMMUNOHISTOCHEMISTRY-PARAFFIN PROTOCOL
- June 19-2018
- IMMUNOFLUORESCENCE PROTOCOL
- September 08-2020
- FLOW-CYTOMEYRT-PROTOCOL
- May 20-2022
- Cell-Based ELISA│解您多样本WB检测之困扰
- July 13-2018
- CELL-BASED-ELISA-PROTOCOL-FOR-ACETYL-PROTEIN
- July 13-2018
- CELL-BASED-ELISA-PROTOCOL-FOR-PHOSPHO-PROTEIN
- July 13-2018
- Antibody-FAQs
- Products Images
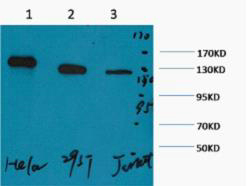
- Western blot analysis of 1) Hela, 2) 293T, 3) Jurkat, diluted at 1:2000. cells nucleus extracted by Minute TM Cytoplasmic and Nuclear Fractionation kit (SC-003,Inventbiotech,MN,USA).
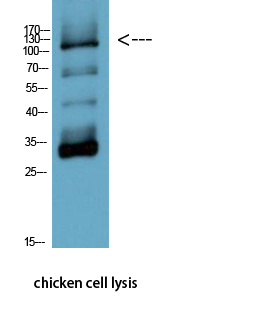
- Western Blot analysis of chicken cell lysis using Antibody diluted at 1:1000

